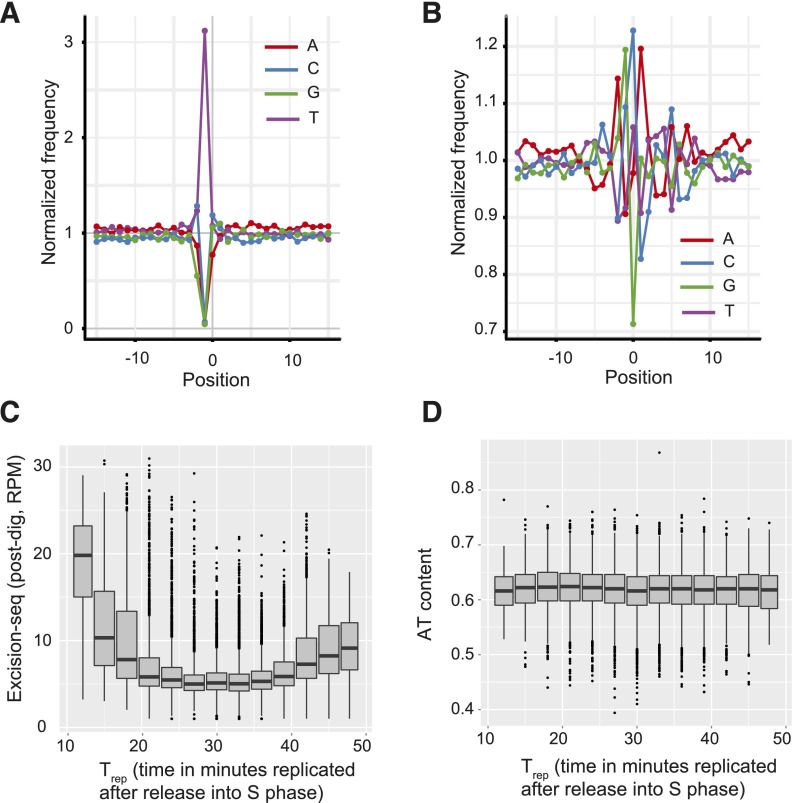
Figure 2.
Excision-seq mapping of uracil content in the budding yeast genome. (A) Normalized frequency of nucleotides relative to mapped positions of sequences from predigestion Excision-seq libraries for mapping S. cerevisiae uracil content. Position 0 corresponds to the mapped position of the 5′ end of 11,326,044 sequencing reads; negative numbers are upstream. Frequencies were normalized to genomic mononucleotide frequencies. (B) Normalized frequency of nucleotides relative to mapped positions of sequences from post-digestion Excision-seq libraries for mapping S. cerevisiae uracil content. Position 0 corresponds to the mapped position of the 5′ end of 2,939,357 sequencing reads (frequency normalization and colors identical to A). (C) Boxplot comparing post-digestion Excision-seq mapping of uracil in S. cerevisiae to replication timing (Trep) (Raghuraman et al. 2001). Mean signals were calculated in 500-bp windows across the genome. (D) Boxplot comparing mean genomic AT content in S. cerevisiae to replication timing; 500-bp regions are identical to C.