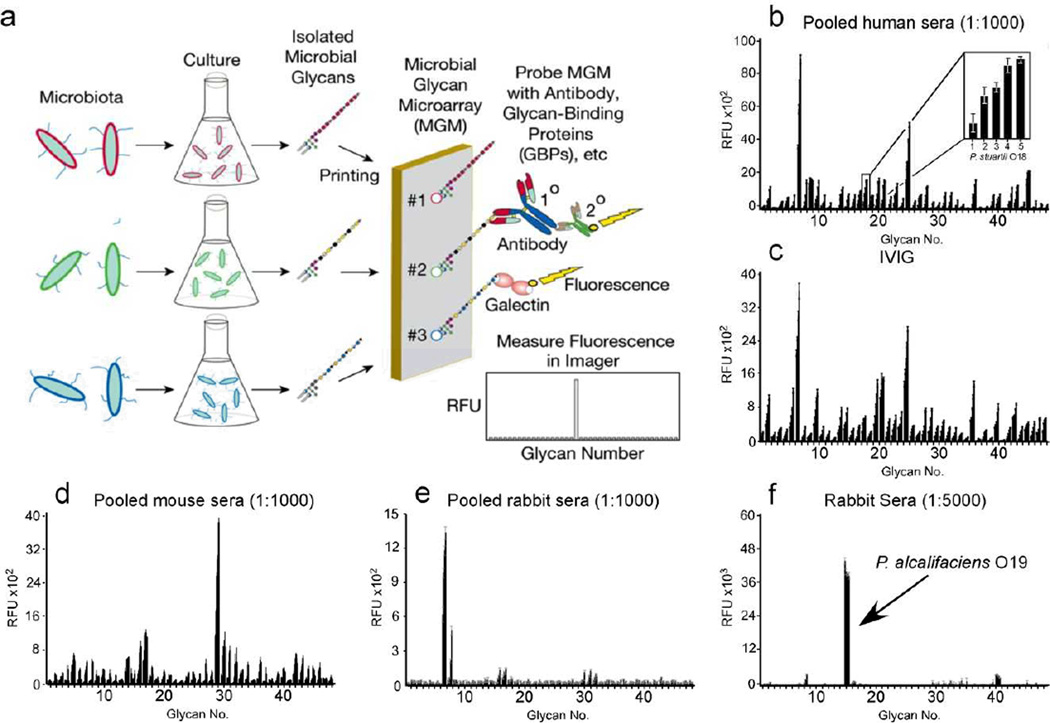
Figure 1. Production and validation of MGM and recognition of microbial glycan structures by sera.
(a) Schematic of MGM design and utility. (b) MGM data obtained after incubation with pooled normal human sera at 1:1000 followed by labeled anti-IgG and anti-IgM). b inset: magnification of antibody interactions with increasing printed concentration of Providencia stuartii O18 BPS (31.25, 62.5, 125, 250, 500 µg/mL). (c) MGM probed with 200 µg/ml IVIG pooled from ~10,000 human donors followed by detection with anti-IgG. (d,e) Detection of pooled normal mouse sera at 1:1000 (d) or pooled normal rabbit sera at 1:1000 (e) with anti-mouse or anti-rabbit IgG, respectively. (f) MGM data obtained after incubation with 1:5000 dilution of sera from rabbits challenged with the indicated bacterial species followed by detection with anti-rabbit IgG. See Supplementary Dataset 1 for complete microarray data.