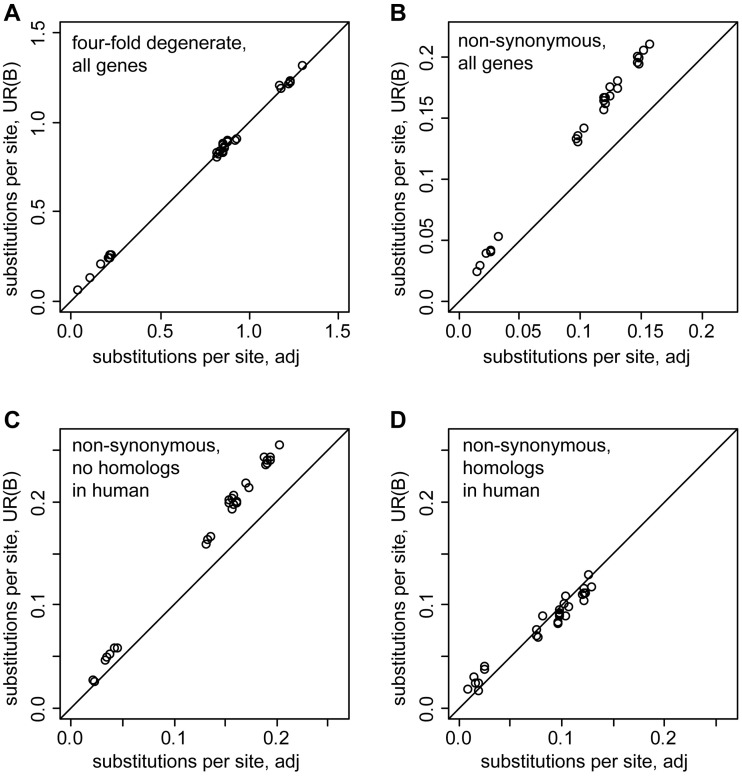
Fig. 1.—
Comparison of phylogenetic distances for genes located in UR(B) and adjacent regions. The ORF alignments for all genes in the indicated groups were merged into a single contig, and the pairwise distances (in substitutions per site) were calculated with MEGA4 using the Pamilo–Bianchi–Li method for all combinations of dm3, droSim1, droYak2, droEre2, droAna3, dp4, droPer1, droWil1 (see Materials and Methods for details). The pairwise distances for UR(B) genes are shown on the ordinate, the distance for the genes in the adjacent regions is on the abscissa. Black lines indicate the expected identical divergence in UR(B) and the adjacent regions. (A) Divergence at 4-fold degenerate sites is essentially identical in UR(B) and the adjacent regions. (B) Genes in UR(B) display more nonsynonymous substitutions than the adjacent genes. (C) Nonsynonymous sites of genes without human homologs show a higher divergence in UR(B) than in the adjacent regions. (D) The divergence at the nonsynonymous sites of the genes with human homologs is very similar in UR(B) and the adjacent regions. Note that the protein sequences of the genes with human homologs evolve slower than the genes without human homologs shown on the panel (C).