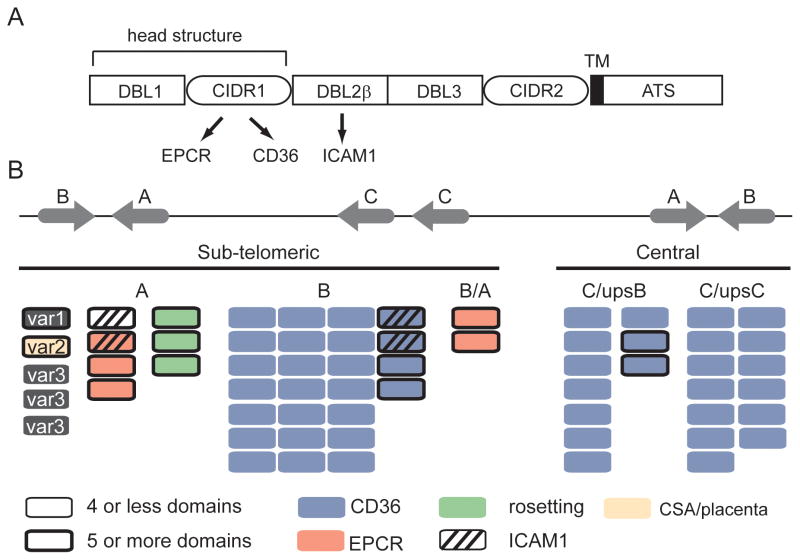
Fig 2.
The genome organization and predicted PfEMP1 binding properties of the 3D7 genome reference isolate. (A) Schematic representation of PfEMP1 domain architecture. The CIDR domain in the PfEMP1 head structure has diverged into CD36 (group B and C) and EPCR (group A) binding variants; binding is highly predictable by adhesion domain classification. ICAM1 binding has been mapped to specific DBLβ subtypes following the PfEMP1 head structure. (B) Group A and B var genes are located in sub-telomeric regions and group C var in central chromosome regions [4]. The predicted binding properties for the 3D7 genome reference isolate are shown based on adhesion domain classification and recombinant protein binding studies. Blue proteins encode CD36-binding CIDR domains [9], red proteins encode EPCR-binding CIDR domains [23], green proteins encode head structures associated with rosetting [27], striped proteins encode DBLβ3 or DBLβ5 domains associated with ICAM1 binding [37–39], and the beige protein (VAR2CSA) is linked to CSA binding in the placenta [7]. Large PfEMP1 proteins with 5 or more domains are distinguished by a thick border.