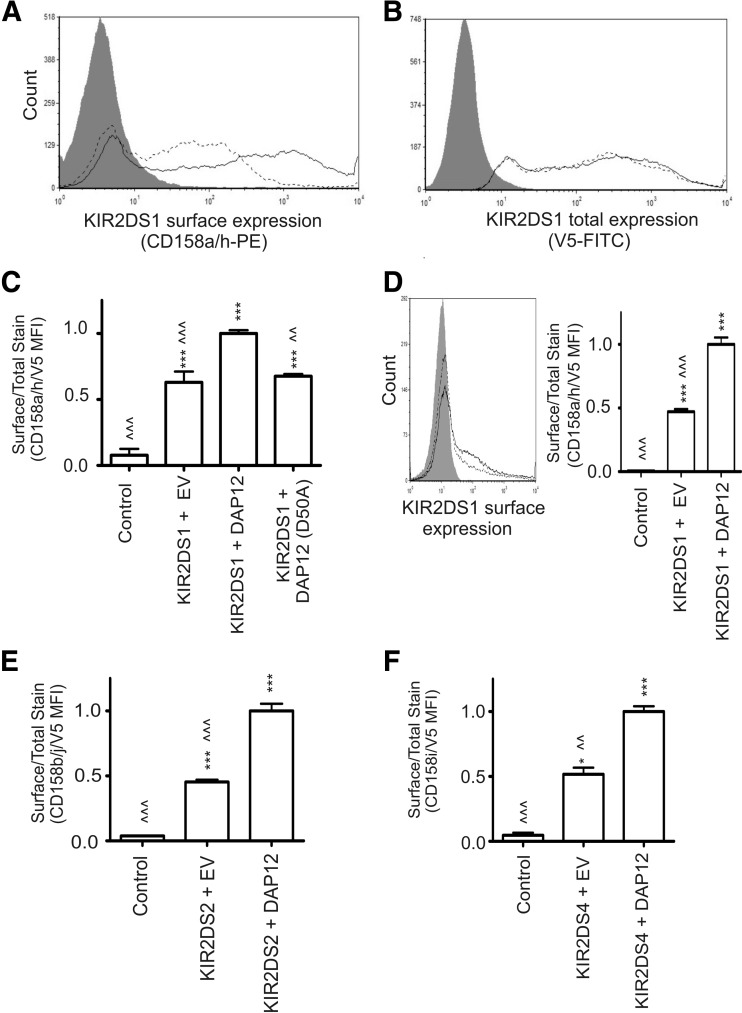
Figure 1. KIR2DS surface expression is increased by exogenous DAP12 expression.
(A) Surface expression of KIR2DS1 on NKL cells following cotransfection of KIR2DS1 with DAP12 (solid line) or empty vector (EV; dashed line). NKL cells transfected with KIR2DS4 (gray shade) that does not interact with the KIR2DS1-specific antibody, was used as a negative control. (B) Total KIR2DS1 expression in NKL cells transfected with empty vector (gray) or NKL cells cotransfected with KIR2DS1 and empty vector (dashed line) or with DAP12 (solid line). Cells were permeablized and probed with a V5-specific antibody for the internal, C-terminal V5 tag linked to each KIR molecule. (C) KIR2DS1 surface expression presented as a ratio of surface-expressed KIR2DS1 (A) to total KIR2DS1 expression (B) following cotransfection with empty vector, WT DAP12, or mutant DAP12 (D50A). All ratios obtained were normalized to results for KIR2DS with exogenous DAP12. (D) KIR2DS1 surface expression on Jurkat cells following cotransfection with DAP12 (solid line), empty vector (dashed line), or transfection with KIR2DS4 (gray shade). The results are presented as a ratio of surface-expressed KIR2DS1 to total KIR2DS1 expression, as described in Materials and Methods. (E and F) Surface-expression data presented in relative ratios for KIR2DS2 and KIR2DS4. KIR2DS1 was used as a negative control for external staining for KIR2DS2 and KIR2DS4, whereas KIR2DS4 was used as the negative control for the KIR2DS1. Each experiment was performed with three independent biological samples, resulting from three independent transfections/condition. The data (mean±sem) represent the results observed in two independent experiments. All experiments were performed in triplicate, and the data (mean±sem) represent the results observed in two independent experiments. Statistical analysis was performed using a one-way ANOVA, followed by Bonferroni's multiple comparison analysis (vs. control: *P<0.05; ***P<0.001 vs. KIR2DS+DAP12: <<P<0.01; <<<P<0.001).