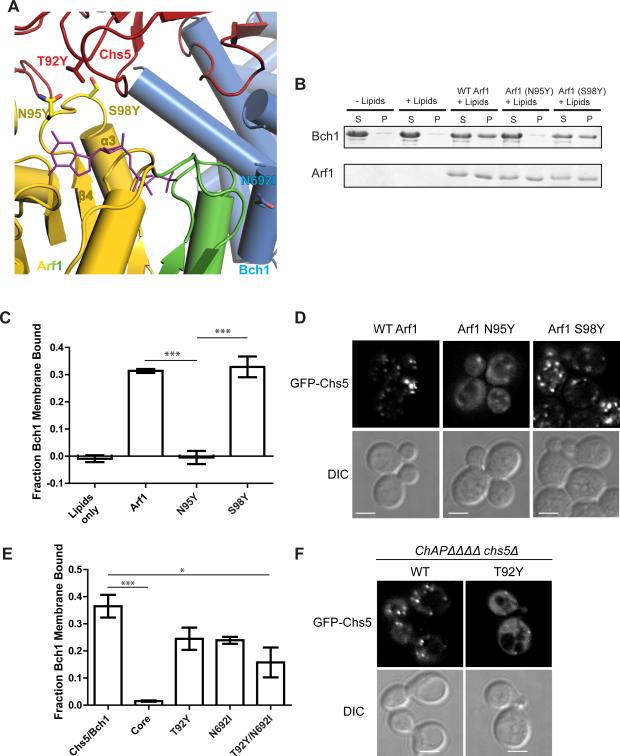
Figure 2. Two interfaces mediate the Arf1-exomer interaction.
A) Close-up view of the two interfaces Arf1 makes with the exomer complex shown in a ribbon diagram. Chs5 is shown in red, Bch1 in blue, and Arf1 in gold, with its switch residues highlighted in green. The nucleotide, GMPPNP, is shown in purple. Mutated residues are shown as sticks.
B) Liposome pelleting assay to assess membrane recruitment of the Chs5(1-299)/Bch1 exomer complex by myristoylated Arf1. N95Y and S98Y are Arf1 mutants. S, supernatant. P, pellet. The appearance of protein in the pellet fraction indicates membrane binding. For simplicity, only the Bch1 bands are shown for exomer complexes. See also Figures S2 and S3.
C) Quantification of the data shown in panel (B). Error bars represent SEM, n = 4, with significance determined by one-way ANOVA with post-processing to correct for multiple comparisons. Only significant comparisons are noted.
D) Images of plasmid-borne GFP-Chs5 in strains harboring wild-type or mutant Arf1. A single deconvolved focal plane is shown at equivalent light levels for each experiment. Scale bar, 2 μm.
E) Quantification of membrane recruitment of Chs5(1-299)/Bch1 mutants. “Core” is the Chs5(1-77)/Bch1 complex which lacks the FBE domain. T92Y is Chs5-T92Y, N692I is Bch1-N692I.
F) Plasmids expressing GFP-Chs5(1-299), wild-type or T92Y mutant, were introduced into a strain lacking exomer (chs5Δ chs6Δ bud7Δ bch1Δ bch2Δ), and imaged. Scale bar 2 μm.