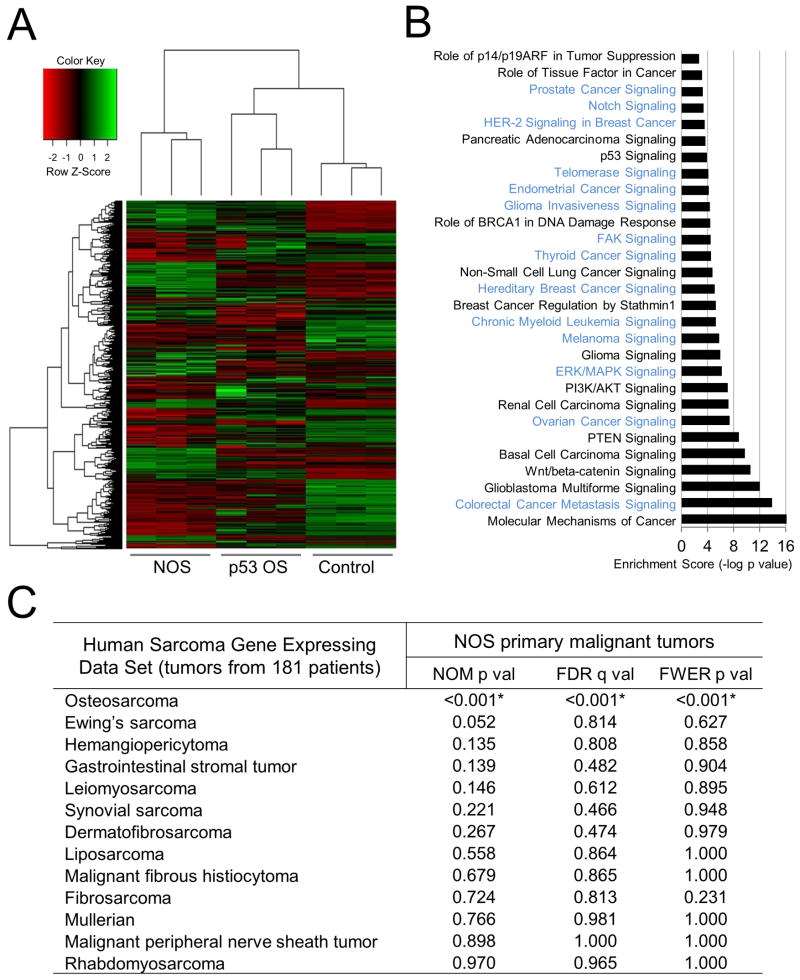
Figure 5. Expression signature of NOS compared to p53-loss-induced OS.
(A) Unsupervised hierarchical clustering of mouse samples and heat map of RNA-sequencing data between Notch OS or p53-loss-induced tumors (p53 OS)
(B) Ingenuity Pathways Analysis (IPA) of cancer canonical signaling pathways associated with significantly regulated genes (p<0.05) in Notch OS versus control. Significantly changed pathways only observed in NOS are colored in light blue, and those changed in both NOS and p53-induced OS are shown in black.
(C) GSEA results for each of the indicated human sarcomas for NOS primary malignant tumors. Asterisks indicate statistical significance. NOM p val, nominal p value; FDR, false discovery rate; FWER, family-wise error rate. See also Figure S5, Tables S1, S2, S3, and S4.