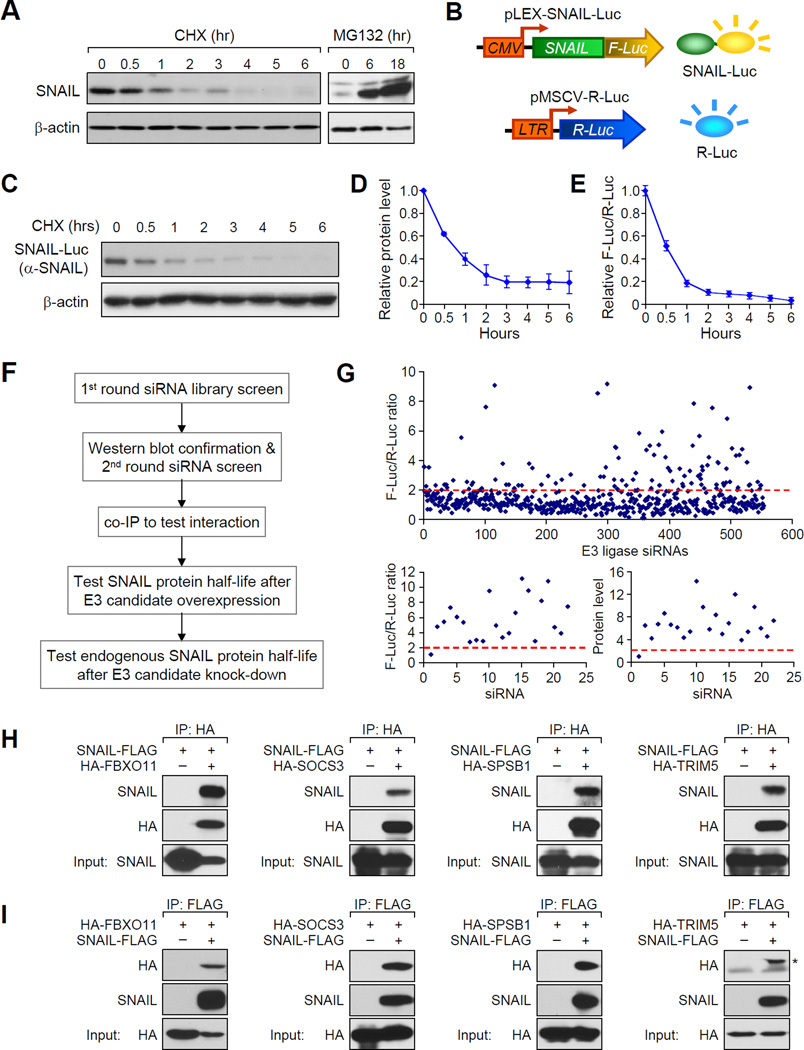
Figure 1. A genome-wide functional screen for E3 ubiquitin ligase(s) targeting SNAIL protein for degradation.
(A) In SUM1315 cells, endogenous SNAIL protein was detected by western blot after CHX (left panel) or MG132 treatment (right panel) for the indicated hours. Western blot data was quantified in Figure S1A. (B) Illustration of dual-luciferase reporter screening system for SNAIL-targeting E3 ligases. (C, D) CHX pulse-chase experiment demonstrated the degradation of SNAIL-F-Luc fusion protein in SUM-SNAIL-Luc/R-Luc cells. Western blot data was quantified in (D). Data is presented as mean ± SEM. (E) Degradation of SNAIL-F-Luc protein was monitored by normalized luciferase activity measurement. Data is presented as mean ± SEM. (F) Experimental procedure flow chart for identification of the E3 ubiquitin ligase(s) targeting SNAIL protein. See text for details. (G) Luciferase based siRNA library screen against human E3 ligases identified multiple E3 candidates, when knocked down in SUM-SNAIL-Luc/R-Luc cells, increased luciferase activity by more than 2 fold (upper panel). A second round of siRNA screening (lower left panel) and immunoblotting (lower right panel) was performed for confirmation of candidates. (H, I) 293T cells were transfected according to each panel labeling. Co-IP experiment was performed using either an HA antibody to pull down HA-tagged E3 ligase proteins (H) or an anti-FLAG antibody against SNAIL-FLAG protein. See also Figure S1 and Table S1 and S2.