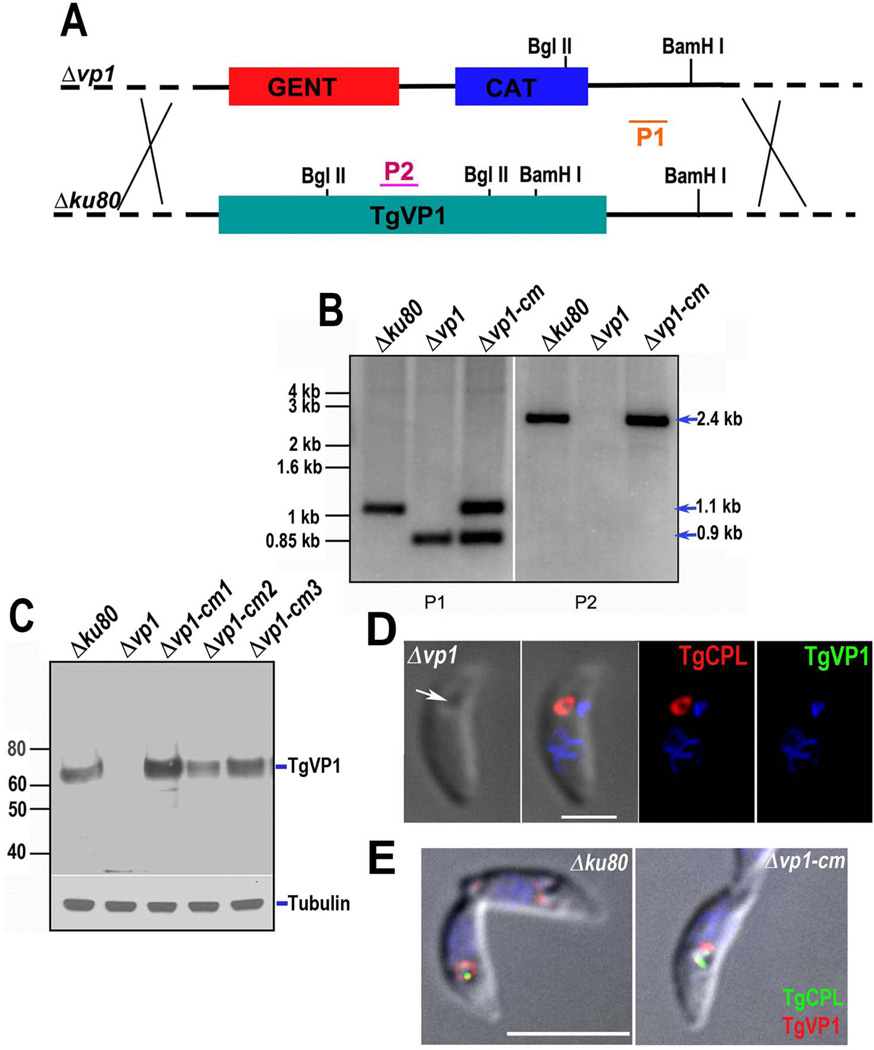
Fig. 1.
Targeted disruption and complementation of the TgVP1 gene. (A) Schematic representation of homologous recombination event in the T. gondii genome for cosmid-based gene disruption. The modified cosmid was transfected into T. gondii tachyzoites (Δku80) following standard procedures and one clone (Δvp1) was selected for further analysis. P1 and P2 indicate probes used for the Southern blot analysis in B. (B) Southern blot analysis of genomic DNA isolated from tachyzoites of Δku80, Δvp1 and Δvp1-cm clones and digested with BamHI and BglII. Probe P1 hybridizes to the 3’ non-coding region present in both loci. P2 hybridizes to a TgVP1 intron only present in the Δku80 and Δvp1-cm clones. (C) Western blot analysis of total tachyzoite lysates using a Guinea pig anti-TgVP1 antibody (1:500). Mouse anti-α-tubulin (1:1,000) was used as loading control. (D) IFA analysis of the Δvp1 clone using mouse anti-CPL (1:1,000) and Guinea pig anti-TgVP1 (1:100). Secondary antibodies were goat-anti-guinea pig (1:1,000) and goat-anti-mouse (1:1,000). The white arrow shows the PLV localization. The PLV is labeled with anti-CPL (red) but shows no signal when using anti-TgVP1 (green) antibody. Scale bar = 2 µm. (E) IFA analysis of the Δku80 and Δvp1-cm strains with mouse anti-CPL (1:200) and rabbit anti-TgVP1 (1:4,000). The PLV is labeled with anti-CPL (green) and anti-TgVP1 (red). Scale bar: 5 µM