Abstract
Human noroviruses (NoVs) are known to recognize histo-blood group antigens (HBGAs) as attachment factors. We report the first experimental evidence that sialic acid-containing glycosphingolipids (gangliosides) are also ligands for human NoVs. Electrospray ionization mass spectrometry-based carbohydrate binding measurements performed on assemblies (P dimer, P particle, and virus-like particle) of recombinant viral capsid proteins of two NoV strains, VA387 (GII.4) and VA115 (GI.3), identified binding to the oligosaccharides of mono-, di-, and trisialylated gangliosides. The intrinsic (per binding site) affinities measured for these ligands are similar in magnitude (102–103 M–1) to those of human HBGAs. Binding of NoV VLPs, P particles, and glutathione S-transferase (GST)-P domain fusion proteins to sialic acid-containing glycoconjugates, observed in enzyme-linked immunosorbent assays, provided additional confirmation of the NoV–ganglioside interactions.
Introduction
Noroviruses (NoVs), a group of small, round-structured RNA viruses constituting the Norovirus genus in the family Caliciviridae, infect both humans and animals. Human NoVs cause epidemic acute gastroenteritis, affecting millions of people and claiming over 200,000 lives annually worldwide.1,2 At present, there is no effective vaccine or antiviral against human NoVs. Structurally, NoVs are nonenveloped, containing an outer protein capsid that encapsulates the single-stranded, positive sense RNA genome of ∼7.7 kb. The NoV capsid is made from a single major structural viral protein, VP1. Crystallography of recombinant NoV-like particles (VLPs) reveals a T = 3 icosahedral symmetry consisting of 180 copies of VP1 organized into 90 dimers.3 VP1 is divided into two major domains, the shell (S) and the protruding (P) domains. The S domain forms the interior, icosahedral shell; while the P domain forms the dimeric protrusions extending outward from the shell.3 The P domain can be further divided into P1 and P2 subdomains, corresponding to the legs and the head of the arch-like protrusion, respectively. The P2 subdomain forms the outermost surface of the capsid with highly variable sequence, responsible for the virus–host interactions and immune recognitions of NoVs.2,4−6
Human NoVs are difficult to study due to the lack of an efficient cell culture system and a small animal model. Currently, research into NoV–host interactions relies on various NoV subviral particles. Expression of full-length VP1 results in VLPs that are structurally similar to an authentic virus.3 Furthermore, expression of various subdomains results in smaller subviral particles or complexes. For example, production of the S domain forms S particles,7,8 corresponding to the interior shell of the capsid, while expressions of the P domains with or without modifications can form P dimers,8−12 12-mer small P particles,13 or 24-mer P particles.14,15 In addition, various glutathione S-transferase (GST)-P domain fusion proteins have been shown to form polyvalent complexes owing to the dimeric and oligomeric features of the GST and the P domain.16,17 These VLPs, P particles and P complexes retain the basic structures of the capsid or P dimer, recognize host ligands and, thus, have been used as tools or models for the study of NoV–host interactions.
Human NoVs recognize histo-blood group antigens (HBGAs) as attachment factors or receptors, which play an important role in the host susceptibility of NoV infection, as shown by both human challenge studies and outbreak investigations.18−20 HBGAs are oligosaccharides linked to membrane proteins or lipids as glycoprotein or glycolipid that are distributed extensively on the surfaces of red blood cells and mucosal epithelia.21 They are also present as free oligosaccharides in biological fluids, such as saliva or milk.21 Human NoVs interact with HBGAs in a strain-specific manner, whereby a number of NoV-HBGA binding patterns involved in all ABO, Lewis and secretor/nonsecretor types have been identified.22,23 The structural basis of these interactions have been elucidated by X-ray crystallography of NoV P dimers in complex with HBGA oligosaccharides.9−12,24 However, it has been observed that some human NoVs, such as VA115 (GI.3),23 Desert Shield virus (GI.3)23 and Noda485 (GII.1),25 do not bind any HBGAs. A human challenge study of Snow Mountain virus (SMV, GII.2) did not reveal a dependence of host susceptibility on HBGA type, despite the fact that the SMV VLP recognizes only the B antigen.26 In addition, a recent study showed that NoV VLPs of Ueno 7k (GII.6) and Noda485 binds Caco-2 cells and human small intestinal epithelium biopsy in a HBGA-independent manner.27 These data suggest that HBGAs may not be the only receptors for human NoVs.
Recent studies have implicated glycosphingolipids and acidic oligosaccharides as human NoV ligands. For example, using thin-layer chromatography and quartz crystal microbalance with dissipation monitoring, Larson et al. reported binding of GII.4 VLPs to galactosylceramide and HBGA glycosphingolipids that were purified from human meconium samples.28,29 Takeda and co-workers demonstrated that VLPs of GII NoVs bound heparan sulfate on the cell surface,30 while Belliot and co-workers showed that GII.4 VLPs recognized sialic acid-containing carbohydrates, such as sialyl Lewis X (LeX), sialyl-lacto-N-fucopentaose, sialyl-lacto-N-tetraose, and sialyl-lacto-N-neotetraose, with affinities comparable to those of HBGA ligands.31 Using saturation transfer difference nuclear magnetic resonance spectroscopy, Peters and co-workers detected the interactions between GII.4 VLPs and the sialic acid moiety of sialyl LeX and sialyl Lea.32 However, they also found that carbohydrates containing sialic acid, but not fucose, e.g. 3′-sialyllactose and 6′-sialyllactose, do not exhibit detectable binding with the VLP.32 The results of these studies, taken together, imply that sialic acid-containing oligosaccharides could also be ligands of human NoVs. In fact, sialic acid-containing oligosaccharides have been shown to be ligands or receptors for some animal caliciviruses (CVs), including murine NoV (MNV1),33 feline calicivirus (FCV)34 and a porcine sapovirus (PSaV, Cowden strain).35 However, solid evidence to establish the ligand status of sialic acid for human CVs (human NoVs and human sapoviruses) is lacking.
Here, we report the first experimental evidence that human NoVs recognize sialic acid-containing glycosphingolipids (gangliosides). The catch-and-release electrospray ionization mass spectrometry (CaR-ESI-MS) assay36 was used to screen a library of gangliosides against the P particle of human NoV VA387 (GII.4). The affinities of 13 gangliosides for the P dimer of VA387 and of a second human NoV strain, VA115 (GI.3), were measured using the direct ESI-MS assay.42 Using a competitive ESI-MS assay, the proxy protein method,38 the highest affinity ligand, GM3, was subjected to additional binding measurements and the affinities for both the VA387 P particle and VLP were determined. Notably, the ganglioside affinities measured for NoV VA387 are comparable to those of known HBGA oligosaccharide receptors.39 Enzyme-linked immunosorbent assays (ELISA) provided additional evidence that both strains of NoVs exhibit binding to sialic acid-containing oligosaccharides.
Experimental Section
Proteins
The VLPs of VA387 (GII.4) were produced in insect cells (SF9) through a recombinant baculovirus containing the gene encoding VA387 VP1 (GenBank accession number AY038600, molecular weight (MW) of monomer 58,887 Da) as described previously.22 The resulting VLPs were purified by sucrose gradient. VA387 P particles (24-mer, MW 865,036 Da), P dimers (MW 69,312 Da), and GST-P domain fusion proteins were produced based on the P domain sequences (residues 222–539) of VP1 via E. coli as reported in our previous studies.16,17 The GST– Gene Fusion System (GE Healthcare Life Sciences, Piscataway, NJ) with plasmid vector pGEX-4T-1 was used for the P proteins expression. Preparations of VA115 (GI.3) VLPs and P particles were attempted based on the VP1 sequences (GenBank accession number AY038598) and the established procedure described above, but the yields for both particles were found to be very low. The P dimers (MW 67,712 Da) and the GST-P fusion proteins of VA115 were produced in high yield (>20 mg L–1 bacteria) through the same procedure as used for the production of the P proteins of VA387. Formations of the 24-mer P particles, P dimers, and the GST-P polymers were analyzed by gel-filtration chromatography via a Superdex 200 size exclusion column (GE Healthcare Life Sciences) controlled by an Akta Fast Performance Liquid Chromatography system (FPLC, Model 920, GE Healthcare Life Sciences).
A single chain fragment (scFv, MW 26,539 Da) of the monoclonal antibody Se155–4 was produced using recombinant technology as described elsewhere.40 A recombinant fragment of the C-terminus of human galectin-3 (Gal-3C, MW 16,330 Da) was generously provided by Prof. C. Cairo (University of Alberta). Bovine ubiquitin (Ubq, MW 8565 Da) was purchased from Sigma-Aldrich Canada (Oakville, Canada). The proteins were concentrated and exchanged into an aqueous 200 mM ammonium acetate (pH 7) using Vivaspin 0.5 mL centrifugal filters (Sartorius Stedim Biotech, Göttingen, Germany) with a MW cutoff of 10 kDa and stored at −80 °C until use. The concentrations of protein stock solutions were estimated by UV absorption.
Ligands
The structures of the oligosaccharides and glycoconjugates used in this study are shown in Figure S1 (Supporting Information). The 17 ganglioside and globoside oligosaccharides (GM3, GM2, GM1a, GM1b, GD3, GD2, GD1a, GD1b, GT3, GT2, GT1a, GT1c, fucosyl-GM1, asialo-GM2, asialo-GM1, Gb3 and Gb4) were purchased from Elicityl SA (Crolles, France). H type 3 trisaccharide, A type 3 tetrasaccharide, and B type 3 tetrasaccharide were a gift from Prof. T. Lowary (University of Alberta).41 Each solid compound was dissolved in ultrafiltered Milli-Q water (Millipore, MA) to give a 1 mM stock solution. The stock solutions were stored at −20 °C until needed. Polyacrylamide (PAA)-conjugated Neu5Ac, 6′-sialylacNAc, and GM3 trisaccharide were purchased from Vector Lab (Burlingame, CA). They were stored at −20 °C until used.
Mass Spectrometry
All of the ESI-MS assays were carried out on a Synapt G2S quadrupole-ion mobility separation-time-of-flight (Q-IMS-TOF) mass spectrometer (Waters, Manchester, U.K.) equipped with a nanoflow ESI (nanoESI) source. The CaR-ESI-MS and direct ESI-MS assays were performed in negative ion mode, whereas the proxy protein ESI-MS assay was implemented in positive ion mode. NanoESI tips were produced from borosilicate capillaries (1.0 mm o.d., 0.68 mm i.d.) pulled to ∼5 μm using a P–1000 micropipette puller (Sutter Instruments, Novato, CA). A platinum wire was inserted into the nanoESI tip, and a capillary voltage was applied to carry out ESI. The source parameters for both negative and positive ion modes were: capillary voltage −0.8 kV (negative ion mode) or 1.0 kV (positive ion mode), source temperature 60 °C, cone voltage 60 V (negative ion mode) or 35 V (positive ion mode), Trap voltage 5 V, and transfer voltage 2 V. Data acquisition and processing were performed using MassLynx software (version 4.1).
CaR-ESI-MS Assay
The CaR-ESI-MS assay was performed to identify carbohydrate ligands of the NoV VA387 P particle. Ions corresponding to ligand-bound P particle were isolated using the quadrupole mass filter. The quadrupole was set to transmit a broad mass-to-charge ratio (m/z) window (approximately 200 m/z units), which allows for the simultaneous passage of free and ligand-bound P particle complexes at a given charge state. Protein–ligand complexes were subjected to collision-induced dissociation (CID) in the Trap region of the Synapt G2S by increasing the Trap voltage from 5 to 200 V. Argon (1.42 × 10–2 mbar) was used to carry out CID in the Trap region. In most instances, the deprotonated ligands released from the complexes could be identified from their MWs. Where required, IMS was used to separate the released isomeric ligands. For IMS separation a wave height of 35 V was used, and the wave velocity was ramped from 2000 to 500 m s–1. In all cases a helium flow rate of 150 mL min–1 and a nitrogen flow rate of 40 mL min–1 were used. The arrival time distributions (ATDs) for the released ligands were compared to reference ATDs, which were measured for the deprotonated carbohydrates produced directly from solution.
Direct ESI-MS Assay
The direct ESI-MS assay was used to quantify the affinities of the carbohydrate ligands for the NoV P dimers of VA387 and VA115. At least four different initial ligand concentrations were used for each oligosaccharide tested, and the binding measurements were carried out in triplicate. A complete description of the data analysis method employed to calculate the intrinsic association constants (Ka,int) can be found elsewhere.42,37 Briefly, the abundance ratio (Ri) of the ligand-bound protein (PLi), bound to i molecules of L, to free protein (P) measured by ESI-MS (after correction for nonspecific ligand-protein binding) is taken to be equal to the equilibrium concentration ratio in solution, eq 1:
| 1 |
Assuming the protein has h independent and identical binding sites, Ka,int can be expressed by eq 2:
| 2 |
where [P]0 and [L]0 are the initial concentrations of the protein and ligand, respectively, and f is the fraction of occupied binding sites, eq 3:
| 3 |
In the case of the P dimer, which has two equivalent binding sites, Ka,int can be found using eq 4:
| 4 |
Proxy Protein ESI-MS Method
The proxy protein ESI-MS assay was used to quantify the affinities of GM3 trisaccharide for NoV VA387 P particle and VLP. A complete description of the data analysis method employed to calculate Ka,int can be found elsewhere.38 Briefly, a proxy protein (Pproxy), which binds to L with a known affinity, is used to monitor the extent of L binding to P. Specifically, in the presence of P, the abundance ratio Rproxy (= [PproxyL]/[Pproxy]) will quantitatively reflect the concentration of L bound to P and Ka,int can be evaluated using eq 5:
 |
5 |
where the initial concentrations of target protein ([P]0), proxy protein ([Pproxy]0) and ligand ([L]0) as well as the association constant for binding of Pproxy to the ligand (Ka,Pproxy) are known; [P]m,0 is the initial concentration of binding sites in the target protein, i.e., [P]m,0 = h × [P]0.
Enzyme-Linked Immunosorbent Assay (ELISA)
PAA-conjugated Neu5Ac, 6′-sialylacNAc and GM3 trisaccharide were dissolved in 1X PBS (pH 7.4). They were diluted and coated on a 96-well microtiter plate at concentration of 2 μg mL–1 and stored at 4 °C overnight. After blocking with 5% nonfat dry milk, NoV VLP, P particle, or GST-P domain fusion proteins as well as GST (negative control) at 50 ng μL–1 were added and incubated for 2 h at 37 °C. The ligand-bound NoV VLP and P proteins were detected by homemade guinea pig hyperimmune serum against VA387 VLP and VA115 P protein (1:3000), respectively, followed by horseradish peroxidase (HRP)-conjugated goat antiguinea pig immunoglobulin G (IgG, 1:3000; ICN, Aurora, OH). Bound GST was detected by a homemade GST antibody. The signals were displayed using a TMB kit (Thermo Fisher Scientific, Rockford, IL).
Results and Discussion
Ganglioside Binding to NoV VA387 P Particle
Evidence of ganglioside binding to NoVs was initially revealed through the screening of a small (20 components) carbohydrate library against the P particle (24-mer, MW 865,036 Da) of NoV VA387 (GII.4) using the CaR-ESI-MS assay.36 The library consisted of the oligosaccharides of 17 glycosphingolipids, GM1a, GM1b, GM2, GM3, GD1a, GD1b, GD2, GD3, GT1a, GT1c, GT2, GT3, fucosyl-GM1 (referred to as Fuc-GM1), asialo GM1, asialo GM2, Gb3 and Gb4 as well as three known HBGA oligosaccharide ligands, H type 3 trisaccharide (referred to as H3), A type 3 tetrasaccharide (A3), and B type 3 tetrasaccharide (B3). The intrinsic affinities of the HBGA ligands range from 700 to 1500 M–1.39 The CaR-ESI-MS assay was carried out by first incubating the P particle with the carbohydrate library, followed by direct ESI-MS analysis of the mixture. Because of the high MW of the P particle, the identity of the bound ligands could not be established directly from the mass spectrum. Instead, using a quadrupole mass filter set to pass a range of mass-to-charge-ratio (m/z) ions, all of the ligand-bound P particle ions at a given charge state were isolated and then activated (heated) using CID to release the ligands (as ions) from the complex. Given that carbohydrates have relatively low gas-phase acidities and are able to effectively compete with proteins for negative charge, the CaR-ESI-MS assay was carried out in negative ion mode.36 Accurate mass analysis, alone or in combination with ion mobility separation (IMS), which separates ions based on size and shape, allowed for positive ligand identification.
Shown in Figure 1a is a representative ESI mass spectrum acquired in negative ion mode for an aqueous ammonium acetate (200 mM, pH 7, 25 °C) solution of P particle (3 μM) and the carbohydrate library (10 μM each). From the mass spectrum it can be seen that the P particle exists predominately as a 24-mer, with a charge state distribution ranging from −60 to −65. Signal corresponding to an 18-mer is also present, although at lower abundance, with a charge state distribution of −51 to −54. Due to the high MW of the P particle and the formation of adducts during the ESI process, it was impossible to resolve the ions corresponding to free P particle and its complexes with one or more oligosaccharide ligands. However, CID, performed using a 200 m/z wide isolation window centered at 14,350 to pass ions corresponding to the −61 charge state of the P particle, led to the appearance of singly deprotonated ions of the three HBGA oligosaccharides as well as GM3 (m/z 632.2), GM2 (m/z 835.3), GD3 (m/z 923.3), GM1a and/or GM1b (m/z 997.3), and Fuc-GM1 (m/z 1143.4) (Figure 1b). Ions corresponding to the singly deprotonated GD2 (m/z 1126.4) and the doubly deprotonated ions of GD1a and/or GD1b (m/z 644.1) were also detected, although at low abundance (Figure 1b). Abundant multiply charged protein monomer ions, Pmn– at n = 10–23, were also evident (Figure 1b). Implementation of the CaR-ESI-MS assay using other charge states of the P particle complexes produced similar results (Figure S2, Supporting Information). Ion mobility separation of the released ligands revealed evidence that both GM1a and GM1b are released from the P particle, with GM1a being more abundant (Figure S3a, Supporting Information). The doubly deprotonated ions of GD1a and GD1b could not be differentiated using optimized IMS conditions (Figure S3b, Supporting Information), and therefore, it was not possible to establish whether one or both oligosaccharides bind to the P particle directly from these measurements. Instead, the CaR-ESI-MS assay was applied to solutions containing P particle (3 μM) and 10 μM of GD1a or GD1b. These data revealed that only GD1a binds to the P particle under these solution conditions (Figure S4, Supporting Information). The CaR-ESI-MS results provide compelling evidence that the P particle of VA387 exhibits a broad specificity for mono- and disialylated gangliosides. However, there is a clear preference for GM3, and the addition of saccharides to Gal (e.g., GM1 or GM2) or Sia (e.g., GD3, GD2 or GD1b) decreases binding, compared to GM3. These data, combined with affinities measured for ganglioside oligosaccharides, vide infra, suggest that the Sia-Gal-Glc moiety represents the dominant recognition epitope for this NoV.
Figure 1.
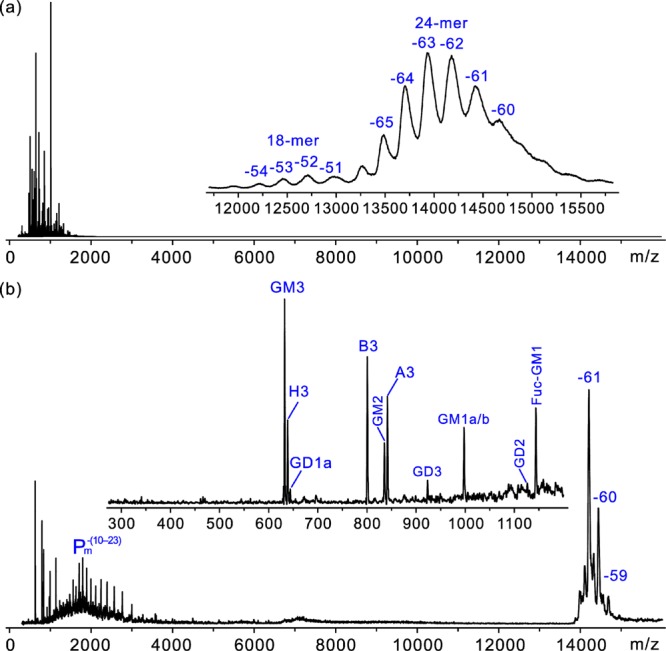
(a) ESI mass spectrum acquired in negative ion mode for an aqueous ammonium acetate solution (200 mM, pH 7 and 25 °C) of NoV VA387 P particle (3 μM) and a 20-component (10 μM each) carbohydrate library consisting of the oligosaccharides of GM1a, GM1b, GM2, GM3, GD1a, GD1b, GD2, GD3, GT1a, GT1c, GT2, GT3, Fuc-GM1, asialo GM1, asialo GM2, Gb3, and Gb4 as well as the H3, B3, and A3 oligosaccharides. (b) CID mass spectrum measured for the −61 charge state of the free and ligand-bound P particle.
Ganglioside Affinities for NoV VA387 Capsid Proteins
Based on the relative abundances of the released oligosaccharide ligands measured by CaR-ESI-MS (Figure 1) it would appear that the affinities of the ganglioside ligands are similar to those of the highest affinity HBGA oligosaccharides.39 However, this conclusion is predicated on the assumption that the release efficiency of the bound-ligands is essentially independent of structure. Because of the presence of the sialic acid, it is possible that gangliosides (which are likely deprotonated in the gaseous complexes) are preferentially released from the P particle due to a lower activation energy resulting from Coulombic repulsion.43,44 Therefore, it was important to measure the affinities directly. In order to do this, the corresponding P dimer of the NoV was used. The affinities were measured using the direct ESI-MS assay, which has been shown to provide reliable Ka values for many protein–carbohydrate interactions.42 Affinities were measured for the oligosaccharides of 13 gangliosides (GM3, GM2, GM1a, GM1b, GD3, GD2, GD1a, GD1b, GT3, GT2, GT1a, GT1c, and Fuc-GM1) for the VA387 P dimer (MW 69,312 Da). A reference protein (Pref) was used in all cases to correct the mass spectra for the occurrence of nonspecific carbohydrate–protein interactions during the ESI process.45,46 A representative ESI mass spectrum acquired for an aqueous ammonium acetate solution (200 mM, pH 7, 25 °C) of VA387 P dimer (12 μM) and GM3 trisaccharide (80 μM) is shown in Figure 2a as well as the distribution of ligand-bound P dimer after correction for nonspecific binding. From the ESI-MS data, Ka,int values were calculated for each oligosaccharide (Table 1). Affinities were also measured for A3, B3, and H3 and shown to agree well with the reported values (Table S1, Supporting Information).39 Inspection of the Ka,int values reveals that, of the tested gangliosides, GM3 exhibits the highest affinity for the VA387 P dimer, which is consistent with the results of the CaR ESI-MS measurements, vide supra. Moreover, the Ka,int (1500 M–1) is identical, within experimental error, to that of B3 (1500 ± 150 M–1).39 Of the 12 other gangliosides investigated, nine bind weakly (Ka,int <500 M–1) and three (GD1b, GT3 and GT1a) do not show any detectable binding. Notably, the quantitative binding data obtained for the P dimer agree qualitatively with the relative affinities inferred from the CaR-ESI-MS measurements performed on the P particle. Moreover, all ligands with affinities >100 M–1 were detected in the CaR-ESI-MS measurements (Table S2, Supporting Information).
Figure 2.
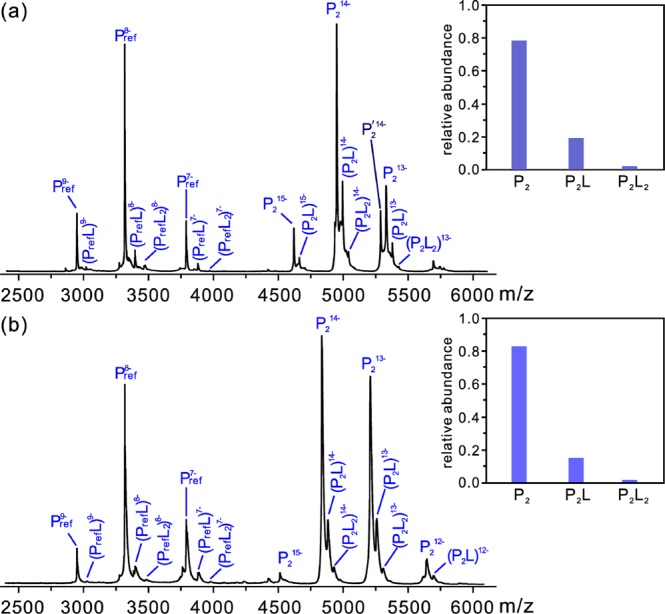
(a) ESI mass spectrum acquired in negative ion mode for aqueous ammonium acetate solution (200 mM, pH 7 and 25 °C) of NoV VA387 P dimer (P2, 12 μM), GM3 trisaccharide (80 μM) and Pref (4 μM). Another minor form of P dimer (P′2, MW 74,080 Da) was also detected with lower abundance. Inset, normalized distribution of GM3 bound to P2 after correction for nonspecific ligand binding. (b) ESI mass spectrum acquired in negative ion mode for aqueous ammonium acetate solution (200 mM, pH 7 and 25 °C) of NoV VA115 P dimer (P2, 12 μM, MW 67,712 Da), GM3 trisaccharide (80 μM), and Pref (4 μM). Inset, normalized distribution of GM3 bound to P2 after correction for nonspecific ligand binding.
Table 1. Intrinsic (per binding site) Association Constants (Ka,int) for P dimer and the Oligosaccharides of 13 Gangliosides Measured in Aqueous Ammonium Acetate (200 mM) at pH 7 and 25 °C Using the Direct ESI-MS Assaya.
| L | Ka,int (M–1) | Ka,int (M–1) |
|---|---|---|
| P dimer VA387 | P dimer VA115 | |
| GM3 | 1500 ± 150 | 1300 ± 130 |
| GM2 | 360 ± 90 | 700 ± 200 |
| GM1a | 350 ± 80 | 400 ± 140 |
| GM1b | 180 ± 60 | 480 ± 110 |
| GD3 | 340 ± 60 | 420 ± 140 |
| GD2 | 150 ± 60 | 700 ± 140 |
| GD1a | <100 | 320 ± 90 |
| GD1b | NBb | 340 ± 50 |
| GT3 | NBb | 310 ± 150 |
| GT2 | <100 | 230 ± 50 |
| GT1a | NBb | 210 ± 70 |
| GT1c | <100 | 260 ± 110 |
| fucosyl-GM1 | 460 ± 150 | 600 ± 130 |
The reported errors are one standard deviation.
NB = No binding detected.
To demonstrate the relevance of the affinity data acquired for the P dimer, affinity measurements were also carried out for GM3 trisaccharide binding to the VA387 P particle and VLP (180-mer, MW ∼10.5 MDa). An adaptation of the proxy protein ESI-MS method, which combines direct ESI-MS binding measurements and competitive protein binding, was used to evaluate the affinities.38 A recombinant fragment of the C-terminus of human galectin-3 (Gal-3C, MW 16,330 Da), which contains a carbohydrate recognition domain and interacts with a β-galactoside moiety,47,48 served as the proxy protein (Pproxy). Importantly, Gal-3C binds to GM3 trisaccharide with an affinity of (1.20 ± 0.02) × 104 M–1. The extent of binding of GM3 trisaccharide to Gal-3C, as determined by ESI-MS, in the presence of known concentrations of the target protein (P particle or VLP) allowed for a quantitative measure of GM3 binding to the target.
ESI-MS measurements were performed on aqueous ammonium acetate solutions (160 mM, pH 7 and 25 °C) of Pproxy (3.0 μM), Pref (1.0 μM), GM3 trisaccharide (40 μM), and either P particle, at concentrations ranging from 0 to 7.2 μM (corresponds to monomer concentration of 0–172.8 μM), or VLP, at concentrations ranging from 0 to 570 nM (monomer concentration of 0–102.6 μM). Representative ESI mass spectra acquired in positive ion mode in the absence and presence of NoV VLP (570 nM) are shown in Figure 3a and 3b, respectively. The distributions of ligand-bound Pproxy, following correction for nonspecific ligand binding, are also given. Inspection of the distributions reveals a measurable decrease in the extent of GM3 trisaccharide binding to Gal-3C upon addition of VLP. This observation confirms that the VLP binds the trisaccharide. The dependence of the extent of GM3 trisaccharide binding to Pproxy on VLP concentration is shown in Figure 3c. Binding measurements performed on solutions containing P particle yielded qualitatively similar results (Figure S5, Supporting Information). Analysis of the Pproxy binding data acquired in the presence of VLP or P particle using the procedure outlined in Experimental Section yields GM3 affinities of 2600 ± 200 and 5500 ± 600 M–1 for the P particle and VLP, respectively. The slight differences in the magnitude of the affinities measured for the binding of a common carbohydrate ligand to the P dimer, P particle, and VLP of a NoV (the first such data set to be reported), likely reflect subtle differences in the structure of the carbohydrate binding site presented by these related protein complexes.3 These differences notwhithstanding, the present results suggest that the P dimer can serve as a surrogate of the VLP for carbohydrate binding studies.
Figure 3.
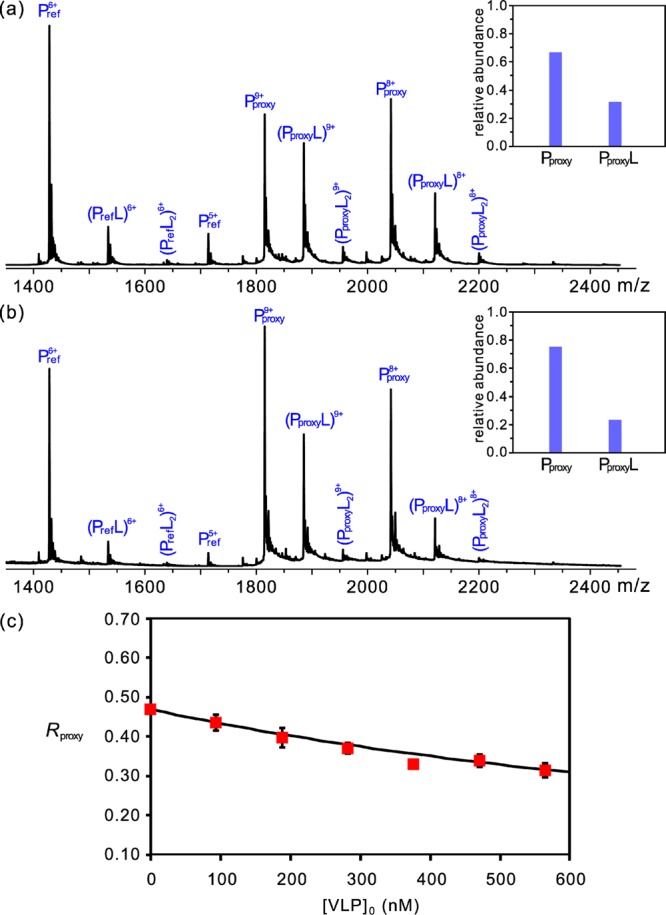
Representative ESI mass spectra measured in positive ion mode for aqueous ammonium acetate solutions (160 mM, pH 7 and 25 °C) of Pproxy (Gal-3C, 3.0 μM), Pref (Ubq, 1.0 μM), and GM3 trisaccharide (40 μM) without (a) or with (b) NoV VA387 VLP (570 nM, 180-mer). Insets show the fraction of free and GM3-bound Pproxy, after correction for nonspecific ligand binding. (c) Plot of the abundance ratio of GM3-bound Pproxy to free Pproxy (Rproxy) versus VLP concentration. The solution conditions for each measurement were the same as in (a), but with the addition of VLP. The curve represents the best fit of eq 5 to the experimental data.
It has been proposed that NoV VA387 has a binding interface that recognizes HBGAs through the α-L-Fuc epitope as the major binding interaction and either the α-D-GalNAc or α-D-Gal epitope as a minor binding interaction.5,9,23 However, these core recognition elements are missing in the ganglioside ligands identified in the present study. Therefore, it is of interest to establish whether the ganglioside ligands interact with the NoV through the HBGA binding site or through a distinct ganglioside binding site. It is not possible to answer this question through competitive binding measurements carried out using a ganglioside oligosaccharide (e.g., GM3 trisaccharide) and VA387 P dimer in the presence of varying concentrations of a HBGA oligosaccharide ligand due to the low affinities of these ligands. Instead, future efforts will rely on X-ray crystallography to establish whether VA387 NoV has distinct binding sites for HBGA and ganglioside ligands.
Ganglioside Affinities for NoV VA115 P Dimer
The aforementioned binding data reveal that NoV VA387 binds to mono- and disialylated gangliosides, with affinities comparable to those of the highest affinity HBGA oligosaccharide ligands. To demonstrate that this is not an isolated example of a human NoV that recognizes gangliosides, the affinities of the 13 ganglioside oligosaccharides for the P dimer of NoV VA115 (GI.3 genotype), which does not bind to human HBGAs,23 were also measured (Table 1). A representative ESI mass spectrum acquired for an aqueous ammonium acetate solution (200 mM, pH 7, 25 °C) of NoV VA115 P dimer (12 μM) and GM3 trisaccharide (80 μM) is shown in Figure 2b as well as the distribution of ligand-bound P dimer after correction for nonspecific binding. Notably, the VA115 P dimer binds to all 13 oligosaccharides tested, and overall, the affinities are slightly higher than those for VA387. These results suggest that human NoVs generally recognize gangliosides as ligands.
Binding of Sialic Acid-Containing Glycoconjugates to NoV VA387 and VA115
Additional evidence for the recognition of sialic acid by human NoVs comes from ELISA measurements carried out on the VA387 VLP, P particle, and GST-P fusion protein16,17 as well as VA115 GST-P fusion protein, with PAA-conjugated Neu5Ac, 6′-sialylacNAc, and GM3 trisaccharide. As shown in Figure 4, the two NoV capsid proteins bind all three sialic acid-containing glycoconjugates. It is curious that the VA387 VLP exhibited weaker binding than that of the P particle to the three glycoconjugates and the cause of the weaker binding is, at this time, unknown. Nevertheless, the fact that all three assemblies of NoV capsid protein exhibit a similar binding pattern to the three glycoconjugates (GM3 > 6′-sialylacNAc > Neu5Ac) validate their applications as models for NoV–ligand interaction. Moreover, comparing the binding of GST-P fusion protein of VA387 to that of VA115 indicates that the sialic acid-containing glycoconjugates have slightly higher affinities for VA115, consistent with the ESI-MS data. These results, together with those from ESI-MS, suggest both α-(2,3)- and α-(2,6)- linked sialic acids as critical motifs in VA387 and VA115 binding, similar to what has been reported for MNV133 and PSaV. 35 It is important to point out that, although sialic acid-containing oligosaccharides have been identified as receptors for an animal NoV (MNV1)33,49 and two other animal CVs (FCV and PSaV),34,35 human NoVs generally recognize gangliosides in addition to HBGAs. Furthermore, human NoVs differ greatly from MNVs in many other important aspects, including host tropism (human vs mouse), clinical manifestation (with vs without diarrhea/vomiting), and pathogenesis.2
Figure 4.
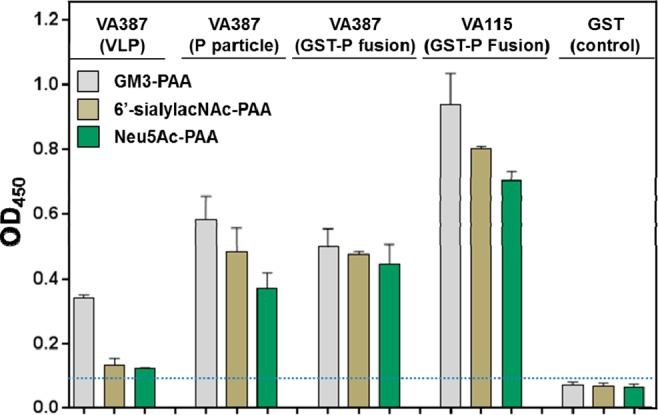
Binding of NoV VLP, P particles, and GST-P fusion protein of VA387 as well as GST-P fusion protein of VA115 to PAA-conjugated GM3 trisaccharide, 6′-sialylacNAc, and Neu5Ac in 1X PBS (pH 7.4). GST, which does not show binding to any of the three glycoconjugates, served as a negative control.
Conclusions
Taken together, the results of ESI-MS and ELISA measurements performed on two human NoVs representing two different genogroups (GI and GII) provide the first experimental evidence of interactions between human NoVs and gangliosides and sialic acid-containing glycoconjugates. Notably, the affinities measured for the oligosaccharides of the ganglioside ligands by ESI-MS are comparable in magnitude to those reported for the oligosaccharides of known HBGA receptors. These experimental data demonstrate sialic acid-containing oligosaccharides as alternative (to HBGAs) ligands for human NoVs and suggest a new mechanism of human NoV–host interaction, one that involves HBGA and sialic acid-containing oligosaccharide receptors and co-receptors for attachment and penetration into host cells and opens a new direction in human NoV research. Further studies to characterize the role of cell surface sialic acids/gangliosides in the early stage of viral infection and its potential coordination with HBGAs for viral attachment and/or entry are needed.
Acknowledgments
The authors are grateful for financial support provided by the Alberta Glycomics Centre and the National Institutes of Health of the Unites States of America and Professors T. Lowary and C. Cairo (University of Alberta) for generously providing oligosaccharides and Gal-3C, respectively, used in this study. L.H. also acknowledges an Alberta Innovates Graduate Student Scholarship.
Supporting Information Available
Carbohydrate structures and mass spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Patel M. M.; Widdowson M. A.; Glass R. I.; Akazawa K.; Vinje J.; Parashar U. D. Emerg. Infect. Dis. 2008, 14, 1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M.; Jiang X. PLoS Pathog. 2010, 6, e1000983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B. V. V; Hardy M. E.; Dokland T.; Bella J.; Rossmann M. G.; Estes M. K. Science 1999, 286, 287. [DOI] [PubMed] [Google Scholar]
- Tan M.; Jiang X. Expert Rev. Mol. Med. 2007, 9, 1. [DOI] [PubMed] [Google Scholar]
- Tan M.; Jiang X. Trends Microbiol. 2011, 19, 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M.; Jiang X. Expert Rev. Mol. Med. 2014, 16, e5. [DOI] [PubMed] [Google Scholar]
- Bertolotti-Ciarlet A.; White L. J.; Chen R.; Prasad B. V. V; Estes M. K. J. Virol. 2002, 76, 4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M.; Hegde R. S.; Jiang X. J. Virol. 2004, 78, 6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S.; Lou Z.; Tan M.; Chen Y.; Liu Y.; Zhang Z.; Zhang X. C.; Jiang X.; Li X.; Rao Z. J. Virol. 2007, 81, 5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. M.; Hutson A. M.; Estes M. K.; Prasad B. V. V Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman G. S.; Biertumpfel C.; Georgiev I.; McLellan J. S.; Chen L.; Zhou T. Q.; Katayama K.; Kwong P. D. J. Virol. 2011, 85, 6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. T.; Tan M.; Xia M.; Hao N.; Zhang X. J. C.; Huang P. W.; Jiang X.; Li X. M.; Rao Z. H. PLoS Pathog. 2011, 7, e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M.; Fang P. A.; Xia M.; Chachiyo T.; Jiang W.; Jiang X. Virology 2011, 410, 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M.; Jiang X. J. Virol. 2005, 79, 14017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M.; Fang P.; Chachiyo T.; Xia M.; Huang P.; Fang Z.; Jiang W.; Jiang X. Virology 2008, 382, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Huang P.; Fang H.; Xia M.; Zhong W.; McNeal M. M.; Jiang X.; Tan M. Biomaterials 2013, 34, 4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Xia M.; Huang P.; Fang H.; Cao D.; Meng X. J.; McNeal M.; Jiang X.; Tan M. Biomaterials 2014, 35, 8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L.; Moe C.; Marionneau S.; Ruvoen N.; Jiang X.; Lindbland L.; Stewart P.; LePendu J.; Baric R. Nat. Med. 2003, 9, 548. [DOI] [PubMed] [Google Scholar]
- Hutson A. M.; Atmar R. L.; Graham D. Y.; Estes M. K. J. Infect. Dis. 2002, 185, 1335. [DOI] [PubMed] [Google Scholar]
- Tan M.; Jin M.; Xie H.; Duan Z.; Jiang X.; Fang Z. J. Med. Virol. 2008, 80, 1296. [DOI] [PubMed] [Google Scholar]
- Ravn V.; Dabelsteen E. APMIS 2000, 108, 1. [DOI] [PubMed] [Google Scholar]
- Huang P.; Farkas T.; Marionneau S.; Zhong W.; Ruvoen-Clouet N.; Morrow A. L.; Altaye M.; Pickering L. K.; Newburg D. S.; LePendu J.; Jiang X. J. Infect. Dis. 2003, 188, 19. [DOI] [PubMed] [Google Scholar]
- Huang P.; Farkas T.; Zhong W.; Thornton S.; Morrow A. L.; Jiang X. J. Virol. 2005, 79, 6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanker S.; Choi J.-M.; Sankaran B.; Atmar R. L.; Estes M. K.; Prasad B. V. V J. Virol. 2011, 85, 8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato H.; Ogawa S.; Ito H.; Sato T.; Kameyama A.; Narimatsu H.; Zheng X. F.; Miyamura T.; Wakita T.; Ishii K.; Takeda N. J. Virol. 2008, 82, 10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L.; Moe C.; LePendu J.; Frelinger J. A.; Treanor J.; Baric R. S. J. Virol. 2005, 79, 2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K.; Kurihara C.; Oka T.; Shimoike T.; Fujii Y.; Takai-Todaka R.; Park Y.; Wakita T.; Matsuda T.; Hokari R.; Miura S.; Katayama K. PLoS One 2013, 8, e66534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydell G. E.; Dahlin A. B.; Hook F.; Larson G. Glycobiology 2009, 19, 1176. [DOI] [PubMed] [Google Scholar]
- Bally M.; Rydell G. E.; Zahn R.; Nasir W.; Eggeling C.; Breimer M. E.; Svensson L.; Hook F.; Larson G. Angew. Chem., Int. Ed. 2012, 51, 12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura M.; Natori K.; Kobayashi M.; Miyamura T.; Takeda N. J. Virol. 2004, 78, 3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rougemont A.; Ruvoen-Clouet N.; Simon B.; Estienney M.; Elie-Caille C.; Aho S.; Pothier P.; Le Pendu J.; Boireau W.; Belliot G. J. Virol. 2011, 85, 4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiege B.; Rademacher C.; Cartmell J.; Kitov P. I.; Parra F.; Peters T. Angew. Chem., Int. Ed. 2012, 51, 928. [DOI] [PubMed] [Google Scholar]
- Taube S.; Perry J. W.; Yetming K.; Patel S. P.; Auble H.; Shu L. M.; Nawar H. F.; Lee C. H.; Connell T. D.; Shayman J. A.; Wobus C. E. J. Virol. 2009, 83, 4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart A. D.; Brown T. D. K. J. Gen. Virol. 2007, 88, 177. [DOI] [PubMed] [Google Scholar]
- Kim D. S.; Hosmillo M.; Alfajaro M. M.; Kim J. Y.; Park J. G.; Son K. Y.; Ryu E. H.; Sorgeloos F.; Kwon H. J.; Park S. J.; Lee W. S.; Cho D.; Kwon J.; Choi J. S.; Kang M. I.; Goodfellow I.; Cho K. O. PLoS Pathog. 2014, 10, e1004172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hawiet A.; Shoemaker G. K.; Daneshfar R.; Kitova E. N.; Klassen J. S. Anal. Chem. 2012, 84, 50. [DOI] [PubMed] [Google Scholar]
- Kitova E. N.; El-Hawiet A.; Schnier P. D.; Klassen J. S. J. Am. Soc. Mass Spectrom. 2012, 23, 431. [DOI] [PubMed] [Google Scholar]
- El-Hawiet A.; Kitova E. N.; Arutyunov D.; Simpson D. J.; Szymanski C. M.; Klassen J. S. Anal. Chem. 2012, 84, 3867. [DOI] [PubMed] [Google Scholar]
- Han L.; Kitov P. I.; Kitova E. N.; Tan M.; Wang L.; Xia M.; Jiang X.; Klassen J. S. Glycobiology 2013, 23, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdanov A.; Li Y.; Bundle D. R.; Deng S. J.; Mackenzie C. R.; Narang S. A.; Young N. M.; Cygler M. Proc. Natl. Acad. Sci. U. S. A. 1994, 91, 6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloncelli P. J.; West L. J.; Lowary T. L. Carbohydr. Res. 2011, 346, 1406. [DOI] [PubMed] [Google Scholar]
- Kitova E. N.; Kitov P. I.; PaszkiewiCz E.; Kim J.; Mulvey G. L.; Armstrong G. D.; Bundle D. R.; Klassen J. S. Glycobiology 2007, 17, 1127. [DOI] [PubMed] [Google Scholar]
- Jiang Y.; Cole R. B. J. Am. Soc. Mass Spectrom. 2005, 16, 60. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Deng L.; Kitova E. N.; Klassen J. S. J. Am. Soc. Mass Spectrom. 2013, 24, 1573. [DOI] [PubMed] [Google Scholar]
- Sun J.; Kitova E. N.; Wang W.; Klassen J. S. Anal. Chem. 2006, 78, 3010. [DOI] [PubMed] [Google Scholar]
- Sun N.; Soya N.; Kitova E. N.; Klassen J. S. J. Am. Soc. Mass Spectrom. 2010, 21, 472. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J.; Hashidate T.; Arata Y.; Nishi N.; Nakamura T.; Hirashima M.; Urashima T.; Oka T.; Futai M.; Muller W. E. G.; Yagi F.; Kasai K. Biochim. Biophys. Acta 2002, 1572, 232. [DOI] [PubMed] [Google Scholar]
- Collins P. M.; Bum-Erdene K.; Yu X.; Blanchard H. J. Mol. Biol. 2014, 426, 1439. [DOI] [PubMed] [Google Scholar]
- Taube S.; Jiang M.; Wobus C. E. Viruses 2010, 2, 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


