Abstract
KAP1/TRIM28/TIF1β was identified nearly twenty years ago as a universal transcriptional co-repressor because it interacts with a large KRAB-containing zinc finger protein (KRAB-ZFP) transcription factor family. Many studies demonstrate that KAP1 affects gene expression by regulating the transcription of KRAB-ZFP-specific loci, trans-repressing as a transcriptional co-repressor or epigenetically modulating chromatin structure. Emerging evidence suggests that KAP1 also functions independent of gene regulation by serving as a SUMO/ubiquitin E3 ligase or signaling scaffold protein to mediate signal transduction. KAP1 is subjected to multiple post-translational modifications (PTMs), including serine/tyrosine phosphorylation, SUMOylation, and acetylation, which coordinately regulate KAP1 function and its protein abundance. KAP1 is involved in multiple aspects of cellular activities, including DNA damage response, virus replication, cytokine production and stem cell pluripotency. Moreover, knockout of KAP1 results in embryonic lethality, indicating that KAP1 is crucial for embryonic development and possibly impacts a wide-range of (patho)physiological manifestations. Indeed, studies from conditional knockout mouse models reveal that KAP1-deficiency significantly impairs vital physiological processes, such as immune maturation, stress vulnerability, hepatic metabolism, gamete development and erythropoiesis. In this review, we summarize and evaluate current literatures involving the biochemical and physiological functions of KAP1. In addition, increasing studies on the clinical relevance of KAP1 in cancer will also be discussed.
Keywords: KRAB domain-associated protein 1, Transcriptional co-repressor, Post-translational modification, Chromatin remodeling, KRAB-containing zinc finger protein
Core tip: This review article primarily summarizes the current findings of KAP1/TRIM28/TIF1β, with focuses on its biochemical and physiological functions. Both the canonical transcriptional co-repressor function and the transcriptional-independent roles of KAP1 are discussed in detail. We highlight the post-translational modifications and the compartmentalized localization of KAP1 and suggest that the function of KAP1 could be spatial and temporal regulated in multiple physiological circumstances. Finally, we summarize the clinical relevance of KAP1 in cancer and discuss the possibility to translate the mechanistic studies of KAP1 to human pathophysiology in the future.
INTRODUCTION
KRAB domain-associated protein 1 (KAP1), also known as tripartite motif-containing 28 (TRIM28) or transcriptional intermediary factor 1 beta (TIF1β) was identified as an interacting protein for Krüppel-associated box zinc finger proteins (KRAB-ZFPs) in 1996[1-4]. Since then, KAP1 has been reported in regulating multiple aspects of physiology, for examples, cell differentiation, DNA damage response (DDR), virus replication, immune response and tumorigenesis (Figure 1). As KAP1 binds to the conserved KRAB repression domain, which is present in many transcription factors, KAP1 is considered as a critical transcriptional co-repressor[2-4]. For instance, many proteins involved in chromatin remodeling or histone modification, such as heterochromatin-associated protein 1 (HP1), nuclear co-repressor (N-CoR), histone deacetylase (HDAC), chromodomain helicase DNA binding protein 3/nucleosome remodeling deacetylase (CHD3/NuRD), histone methyltransferases (HMTs), have been identified in KAP1-containing complexes[5-9] (Table 1). Consequently, KAP1 epigenetically regulates gene expression through multiple transcriptional co-repressor complexes. In addition to regulating KRAB-ZFPs, the activity of transcription factors lacking KRAB domain, such as c-Myc and E2F1, can also be regulated by KAP1[10-14]. KAP1 is subjected to multiple post-translational modifications (PTMs), including phosphorylation and SUMOylation (Figure 2). We and others have demonstrated that these PTMs coordinately regulate the gene repressive function of KAP1[15-18].
Figure 1.
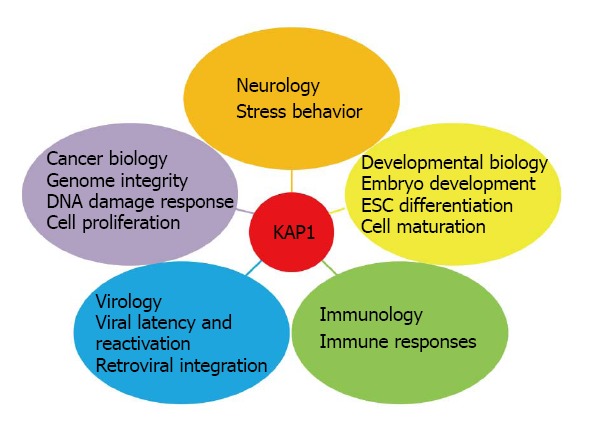
KRAB domain-associated protein 1 is involved in multiple aspects of cellular physiology. Neurology: Kap1 knockout in mouse forebrain induces higher level of anxiety-like behavior. Developmental Biology: Several conditional kap1 deletions impair normal cell development including embryonic stem cell (ESC) differentiation, spermatogenesis, erythropoiesis, and the development of T-cell and B-cell. Immunology: KAP1 is involved in immune responses by regulating T/B cell activity and immune tolerance. Virology: KAP1 is critical to suppress retroviral activation and prevent HIV integration. Cancer Biology: KAP1 is positively or negatively correlated with prognosis in different cancer types. The roles of KAP1 in maintaining genome stability, mediating DNA damage response and affecting cell proliferation in vitro imply its potential roles during tumorigenesis.
Table 1.
Chromatin-associated factors/chromatin-remodeling enzymes interacting with KAP1
| Chromatin-associated factors/chromatin-remodeling enzymes | Consequences of binding with KAP1 | Ref. |
| HP1 | HP1-KAP1 interaction leads to transcriptional repression and has an essential role in development and cell differentiation. Phosphorylation at Ser-473 or Tyr-449, 458, 517 of KAP1 inhibits its interaction with HP1. | [5,6,33,43,46,53,94-97,105] |
| SETDB1 | KAP1 binds to SETDB1 through SUMO:SIM interaction to methylate H3K9 at gene regulatory regions to achieve gene silencing. | [9,15,16,109] |
| N-CoR | N-CoR represses basal transcription by the recruitment of HDACs to deacetylate histones. KAP1 is involved in N-CoR-1 complex to mediate transcriptional repression. | [7] |
| CHD3 (Mi-2α)/NuRD | NuRD complex mediates chromatin remodelling and histone deacetylation via CHD3 (Mi-2α) and HDACs, respectively. KAP1 interacts with NuRD complex via PHD and bromodomain to alter the chromatin structure. | [8,21,37] |
| HDAC1 | KAP1-HDAC1 complex interaction not only regulates histone modification but also non-histone protein deacetylation to exert a variety of different functions (also shown in Table 2). | [113] |
| SMARCAD1 | SMARCAD1 mediates histone deacetylation and associates with KAP1-HDAC1 complex to regulate chromatin marks. | [58] |
| DNMT | KAP1 associates with DNMT to maintain DNA methylation at imprinting control region (also shown in Table 2). | [59,61] |
HP1: Heterochromatin-associated protein 1; ATM: Ataxia-telangiectasia mutated; PP1: Protein phosphatase 1; CHD3/NuRD: Chromodomain helicase DNA binding protein 3/nucleosome remodeling deacetylase; HDAC: Histone deacetylase; N-CoR: Nuclear co-repressor; SETDB1: Bifurcated 1; DNMT: DNA methyltransferase.
Figure 2.
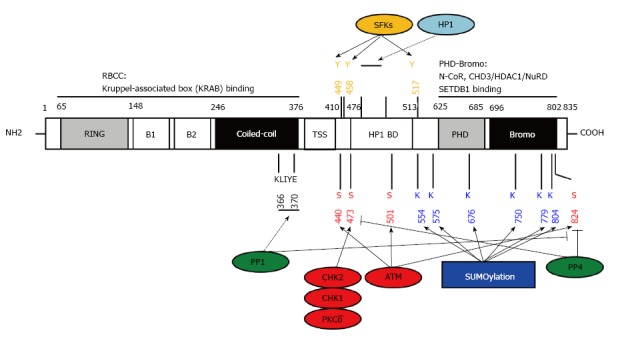
KAP1 structure, post-translational modifications and interacting proteins. KAP1 has multi-domains for protein-protein interaction and post-translational modification. RBCC: RING-B1-B2-coiled-coil; TSS: TIF signature sequence; HP1 BD: HP1 binding domain; PHD: Plant homeo domain. Numbers represent the sequence of amino acids; Blue: SUMOylation sites; Red: Serine phosphorylation sites targeted by the indicated kinases (shown in red) or antagonized by phosphatases (shown in green); Orange: Tyrosine phosphorylation sites targeted by the indicated kinase family; KLIYF: PP1 binding site; PxVxL: HP1 binding site; Dotted lines: Protein-protein interaction; SFKs: Src family kinases; HP1: Heterochromatin-associated protein 1; ATM: Ataxia-telangiectasia mutated; PP1: Protein phosphatase 1; CHD3/NuRD: Chromodomain helicase DNA binding protein 3/nucleosome remodeling deacetylase; HDAC: Histone deacetylase.
Deletion of Kap1 in mouse embryo leads to embryonic lethality[19], suggesting that KAP1 is critical during embryonic development and should be involved in a wide-range of biological/physiological processes. Although most of the KAP1 studies have been focusing on its transcriptional functions, emerging evidence suggests that KAP1 also exerts transcription-independent function. Given that KAP1 functions as a scaffold protein to constitute KAP1-containing complexes that regulate chromatin structure, it also plays an important role in maintaining genome stability by facilitating DNA repair in response to DNA damage through chromatin remodeling[20-22]. Moreover, the RING and the plant homeodomain (PHD) domains of KAP1 possess intrinsic enzymatic activity to potentially catalyze SUMOylation and ubiquitylation[15,23-25]. Interestingly, KAP1 resides in distinct cellular compartments, including the pericentric and centromeric heterochromatin, euchromatin and cytoplasm[6,25,26], implicating its essential functions for different cellular activities. Taken together, the diverse function and regulated subcellular localization suggest that KAP1 impacts multiple aspects of biological processes and it warrants more rigorous investigations of this important protein in the future. In this review, we aim to discuss both the transcriptional and non-transcriptional functions of KAP1. The PTMs of KAP1 involved in these processes will be highlighted. Lastly, the clinical relevance of KAP1 in cancer will also be elaborated.
PROTEIN STRUCTURE AND PTMS OF KAP1
Given that the overall structure of KAP1 has been extensively reviewed previously[27] (and references therein) we will mainly focus on the PTMs of KAP1 and how these PTMs crosstalk to each other and affect the interaction between KAP1 and its partners. KAP1 is a member of TIF1 family, which includes four proteins, TIF1α, TIF1β, TIF1γ and TIF1δ. As the other members in the TIF1 family, KAP1 has an N-terminal tripartite motif (TRIM), RBCC domain, which is composed by a RING finger, 2 B-box zinc fingers and a coiled-coil region. In addition, KAP1 also shares a central TIF1 signature sequence (TSS), an HP1 binding domain (HP1BD), a C-terminal combination of PHD and bromodomain with the other TIF1 members. Different from the other TIF1 proteins, KAP1 does not have a nuclear receptor (NR) box[27] (Figure 2).
The RBCC domain of KAP1 interacts with various KRAB-ZFPs and is considered as an important region for the KAP1 recruitment to KRAB-ZFP binding sites across the genome[28,29]. It has been demonstrated that RBCC domain forms a homotrimer with a single KRAB domain[30]. Interestingly, a recent study suggests that KAP1 can still bind to promoter regions without RBCC domain, suggesting additional mechanisms that might contribute to the KAP1 recruitment to transcription factors on the promoter regions[31]. The TSS domain is adjacent to the RBCC domain, and is required for the transcriptional repressive activity of TIF1γ[32]. However, the function of TSS in KAP1 has yet to been defined.
The hydrophobic PxVxL pentapeptide is located at the central region of KAP1, namely HP1BD. HP1BD interacts with the chromoshadow domain of HP1 proteins and this KAP1-HP1 interaction is critical for the KAP1-mediated gene silencing[5,6,33,34]. It is believed that KAP1-HP1 complex plays a critical role in heterochromatin maintenance and gene silencing. In fact, HP1BD is required for nuclear retention of KAP1 and disrupting KAP1-HP1 interaction reactivates the imprinted gene expression by perturbing histone and DNA methylation levels in vivo[31,35]. However, the detailed mechanism underlying the KAP1 recruitment to HP1 on heterochromatin remains to be elucidated.
The C-terminal PHD and bromodomain of KAP1 (PB domain) recognize histone tail and are also required for the KAP1-mediated gene silencing by recruiting histone modifiers. Specifically, KAP1 interacts with CHD3/NuRD complex, and histone methyltransferase, SET domain, bifurcated 1 (SETDB1)[8,9]. These observations suggest a model for KAP1-dependent recruitment of histone modifiers for histone methylation and heterochromatin formation to achieve gene silencing[27].
Notably, the carboxyl-terminus of KAP1 is subjected to multiple types of PTM. Several studies have revealed that KAP1 is SUMOylated and the SUMOylation of KAP1 is required for its repressive function[15,16,18]. Within or adjacent to the PB domain, six lysines of 554, 575, 676, 750, 779 and 804 have been validated as SUMOylation sites, and the distinct SUMOylation combinations differentially affect the interaction between the bromodomain with SETDB1 and CHD3[15,16,18,28]. KAP1 also undergoes auto-SUMOylation[15]. The magnitude of KAP1 SUMOylation is balanced by deSUMOylases, sentrin specific peptidase 1 (SENP1), SENP7 and the phosphorylation status at serine-824 of KAP1[15,17,36,37].
The serine-824 of KAP1 is primarily phosphorylated by phosphoinositide 3 kinase-like protein kinases (PIKKs), including ataxia-telangiectasia mutated (ATM), ataxia telangiectasia and Rad3 related (ATR), and DNA-dependent Protein Kinase catalytic subunit (DNA-PKcs)[38]. This specific serine 824-phosphorylation is crucial for DDR in different aspects, for examples, the ATM-mediated KAP1 serine-824 phosphorylation is responsible for activating DNA damage checkpoints and chromatin relaxation[17,20], whereas ATR or DNA-PKcs presumably compensates for ATM-deficiency during DDR[39]. Our laboratory has demonstrated that protein phosphatase 1 (PP1) interacts with KAP1 through the PP1-binding motif in the coiled-coil region of KAP1 to dephosphorylate KAP1 at serine-824[36]. In addition, protein phosphatase 4 (PP4) also mediates the KAP1 dephosphorylation at serine-824 and another phosphorylation site at serine-473 upon DNA damage[40,41].
Many DNA damage-inducing agents render both KAP1 serine-824 phosphorylation and serine-473 phosphorylation through ATM-Chk2 or ATR-Chk1 pathways[42]. KAP1 serine-473 phosphorylation is also involved in efficient DNA repair and cell survival upon DNA damage[42-44]. However, unlike the phosphorylation at serine-824, the KAP1 serine-473 phosphorylation is diffusely localized in the nucleus instead of accumulating at damage sites and forming foci[44,45]. It remains to be established whether different KAP1 phosphorylation sites play distinct roles in response to DNA damage. Interestingly, the KAP1 serine-473 phosphorylation regulates cell cycle progression. A study demonstrated that PKCδ phosphorylates KAP1 at serine-473 during S phase of cell cycle. This event dampens the KAP1 and HP1γ interaction and de-represses cyclin A2 to promote S phase progression[46]. Conceivably, KAP1 serine-473 phosphorylation perturbs its association with HP1γ, thereby rendering pan-nuclear distribution. This may be due to the structural alteration of PxVxL domain, which is the HP1 binding site of KAP1. KAP1 mutant that loses its binding ability with HP1 indeed translocates to cytoplasm[31]. This implies that KAP1 serine-473 phosphorylation could function beyond gene regulation in cytoplasm. Results from several studies also suggest that KAP1 serine-473 phosphorylation is associated with immune response. First, KAP1 is constitutively phosphorylated at serine-473 but not serine-824 upon T cell receptor activation in thymocytes[47,48]. Second, Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) infection induces phosphorylation at KAP1 serine-473 in endothelial cells[49]. Many KAP1’s partners, such as STAT1/3, NF-κB and IRF5/7[24,49-52], are involved in inflammation and immune response. Further exploring the role of KAP1 serine-473 phosphorylation in the immune responses induced by viral infection or other types of stress could be rewarding. More recently, Kubota et al[53] reported that tyrosines-449, 458 and 517 of KAP1 are phosphorylated by Src family kinases (SFKs). The phosphorylation of these tyrosine residues also interferes the interaction between KAP1 and HP1. As SFKs are involved in regulating a wide-range of oncogenic processes, including cell growth and differentiation, this finding implies that KAP1 plays a role in SFK-mediated oncogenic transformation.
KAP1 is also demonstrated to be acetylated and the level of KAP1 acetylation is downregulated by HDAC10[54]. Although HDAC10 regulates KAP1 transcriptional co-repressor activity, there is no direct evidence showing whether the acetylation affects KAP1-mediated transcriptional control. Studies on the regulation of KAP1 acetylation and whether KAP1 acetylation crosstalks with other types of KAP1 lysine-PTM are expected to provide additional insights into the roles of PTMs in governing KAP1 function.
TRANSCRIPTIONAL FUNCTIONS OF KAP1
KAP1 was originally identified to be associated with KRAB-ZFPs, suggesting a role of KAP1 in transcriptional control. In addition to the RBCC domain, there are also other domains allowing KAP1 to interact with a wide variety of proteins such as histone acetylases (HATs), HDACs, HMTs and DNA methyltransferases (DNMTs). Thus, KAP1 has the flexibility to regulate transcription through multiple mechanisms, possibly depending on its PTMs, location and specific binding partners (Figure 2, Tables 1 and 2).
Table 2.
Transcription factors involved in KAP1-regulated gene expression
| Transcription factor | KAP1 function | Ref. |
| Myc | KAP1 is involved in MM-1 and HDAC1 complex to suppress c-Myc transcription activity | [10,11] |
| ZNF160 | KAP1 interacts with ZNF160 and recruits HDAC to downregulate TLR4 in intestinal epithelial cells | [79] |
| Oct3/4 | KAP1 is potentially involved in regulating pluripotency of embryonic stem cells | [78] |
| E2F1 | KAP1 suppresses E2F1 transcriptional activity and inhibits E2F1 acetylation in an HDAC1 dependent manner | [13] |
| p53 | KAP1 is associated with MDM2-p53-HDAC1 complex and inhibits p53 acetylation and promotes p53 degradation | [75] |
| p53 | MAGE proteins stabilize KAP1-p53 complex to decrease acetylation and promote degradation of p53 | [80] |
| p53 | KAP1 ubiquitinates p53 for proteasome degradation | [23] |
| IRF7 | KAP1 sumolyates IRF7 and suppresses IRF7 transcriptional activity in IFN production | [24] |
| ZBRK1 | KAP1 interacts with ZBRK1 to repress Gadd45α and p21 gene expression | [16,17,69] |
| HIF-1α | VHLaK potentially recruits KAP1 to HIF-1α complex to suppress HIF-1α downstream gene expression | [72] |
| Nrf2 | KAP1 interacts with Nrf2 and facilitates Nrf2 transactivation activity | [76] |
| STAT3 | KAP1 interacts with STAT3 to suppress STAT3 transcriptional activity | [82] |
| STAT1 | KAP1 interacts with STAT1 to suppress STAT1 transcriptional activity | [50] |
| ZNF689 | ZNF689 potentially recruits KAP1 to suppress autophagy-gene-targeting miRNAs | [71] |
| FOXP3 | KAP1 is recruited by FIK in FOXP3-FIK-KAP1 complex to suppress FOXP3-target genes | [74] |
| NFκB | KAP1 is associated with NFκB and negatively regulates it acetylation and transcriptional activity | [51] |
| SRY | KAP1 is recruited by KRAB-O to SRY binding sites for gene regulation | [68,73] |
| ZFP57 | ZFP57 and KAP1 are associated with NP95-DNMT complex for maintaining DNA methylation at imprinting control region | [59,61,62] |
VHLaK: VHL-associated KRAB-A domain-containing protein; HDAC: Histone deacetylase; DNMT: DNA methyltransferase.
Histone modification and chromatin remodeling
RBCC domain is required for the recruitment of KAP1 to KRAB-ZFPs, and KAP1-binding sites are enriched in the promoter regions of KRAB-ZFPs, suggesting an auto-regulation between KAP1 and KRAB-ZFPs[55]. However, a recent genome-wide study reports that KAP1 does not regulate the expression of those ZFPs and other genes that KAP1 is bound to their promoter regions. Instead, KAP1 regulates the expression of genes with KAP1 binding sites distant from their transcription start sites. Furthermore, deletion of the RBCC domain did not profoundly affect the binding of KAP1 to corresponding promoter regions[31]. Considering that KAP1 interacts with HP1 and other histone modifiers and is involved in long-range transcriptional repression[56], KAP1-mediated chromatin remodeling might contribute to the majority of the KAP1-mediated transcriptional repression.
In addition to the basal transcription, KAP1-mediated chromatin remodeling also participates in the activated transcription. We have shown that the DNA damage-induced KAP1 serine-824 phosphorylation decreases SUMOylated KAP1 to reduce the H3K9 di- and trimethylation, a mark enriched at repressed genes, and to increase H3K14 acetylation, a mark enriched at the active genes, on the promoter regions of KAP1-targeted genes, thereby relaxing the chromatin structure and activating the transcription of pro-arrest and pro-apoptotic genes, including p21, Gadd45α, Bax, Noxa and Puma[16,17,36]. Likewise, the phosphorylation of KAP1 serine-824 by the viral protein kinase induces chromatin remodeling to activate lytic genes to support KSHV lytic replication[57]. Notably, KAP1 mediates the chromatin remodeling within the promoter regions, but not the distal regions of its targeted genes upon DNA damage and viral reactivation[16,57]. It would be interesting to investigate how KAP1 mediates chromatin remodeling at distinct gene regulatory regions during basal and activated transcription. Furthermore, a study showing that KAP1 interacts with a SWI/SNIF-like, ATP-dependent chromatin remodeling protein, SMARCAD1, demonstrates that the KAP1-SMARCAD1 complex regulates global H3K9 trimethylation and H3/H4 deacetylation to maintain the silenced loci during DNA replication[58]. Whether this complex is also responsible for KAP1-mediated transcriptional repression during other circumstances remains unknown.
DNA methylation
Emerging evidence suggests that KAP1-containing complex is not only associated with histone modification, but also regulates DNA methylation. KAP1-mediated DNA methylation is important for genomic imprinting and epigenetic reprogramming during embryogenesis[59-63]. Studies have demonstrated that ZFP57, a KRAB-ZFP important for maintaining DNA methylation at the imprinted loci, recruits KAP1 to imprinting control regions (ICRs) to interact with NP95, a protein responsible for recruiting DNMTs to hemimethylated DNA, resulting in the maintenance of genomic imprinting at ICRs[59,61]. Although these results implicate that KAP1 might indirectly recruit DNMT to specific loci, it remains elusive which domain of KAP1 is responsible for the DNMT recruitment. Histone modification and DNA methylation are inter-dependent, and histone lysine methylation is involved in maintaining genomic imprinting at ICRs[64,65]. Whether KAP1 plays a role in the crosstalk between these two epigenetic regulation pathways warrants further investigation.
Transcriptional co-regulator
Although it is known that KAP1 can regulate gene expression by chromatin remodeling and DNA methylation as discussed in the previous sections, the specificity of KAP1-regulated genes under certain conditions is still largely unknown. Here, we reviewed how KAP1 regulates transcription factors on specific gene loci for gene regulation (Table 2).
KAP1 has no DNA binding domain but is recruited by a variety of different KRAB-ZFPs via its RBCC domain[29,66,67]. Therefore, KAP1 is believed to exert its transcriptional repression by associating with KRAB-ZFPs to specific promoter region of its regulating gene[29]. Although KRAB-ZFP is a large family containing over 400 genes encoding more than 700 predicted proteins, only a few studies directly address the role of KAP1 in regulating the KRAB-ZFP-mediated transcriptional repression[68]. In 1996, KAP1 was for the first time demonstrated to interact with KRAB domain of a human ZFP KOX1/ZNF10 to exert transcriptional repression[4]. Later, ZBRK1 was identified as a sequence specific KRAB-ZFP to recruit KAP1 to suppress Gadd45α and p21 expression[16,17,69]. Our laboratory has demonstrated that KAP1 SUMOylation status dictates the ZBRK1-KAP1-mediated gene repressive function by enhancing histone methylation without altering KAP1-ZBRK1 interaction[16,17]. Another KRAB-ZFP, ZNF160 has been shown to suppress TLR4 through the binding with KAP1 in intestinal epithelial cells for immune tolerance to commensal bacteria in gut[70]. More recently, ZNF689 is identified as a potential KRAB-ZFP for recruiting KAP1 to suppress autophagy-gene-targeting microRNAs (miRNAs). The KAP1-mediated repression of these autophagy-gene-targeting miRNA promotes mitophagy, a specific type of autophagy targeting mitochondria, during erythrocyte maturation[71].
Interestingly, proteins with KRAB domain but not DNA binding domain are also reported to being capable of serving as a bridge for KAP1-mediated gene repression[68,72-74]. The KRAB Only (KRAB-O) protein is one example to recruit KAP1 to the sex determination transcription factor SRY for regulating SRY-targeted genes[68,73]. VHL-associated KRAB-A domain-containing protein (VHLaK) promotes the formation of HIF-1α-VHLaK-KAP1 complex and potentially suppresses HIF-1α signaling[72]. FOXP3-interacting KRAB domain-containing protein (FIK) is another bridge protein to recruit KAP1 to the FOXP3 binding sites for suppressing FOXP3-target genes[74]. Other transcription factors without KRAB domain including Myc, Oct3/4, E2F1, p53, IRF5/7, Nrf2, STAT1/3 and NF-κB can also interact with KAP1 for gene regulation[10,13,24,49-52,75-78]. More investigation is required to understand whether the formation of these KAP1-transcription factor complexes relies on other bridge proteins.
Several studies have reported that KAP1 is capable of modulating the acetylation status of histones or transcription factors. In general, KAP1-mediated HDAC recruitment negatively regulates gene expression. While mainly serving for histone deacetylation and for heterochromatin maintenance[79], the KAP1-HDAC complex is also postulated to negatively regulate the acetylation level of the transcription factors. For example, KAP1 stimulates E2F1-HDAC1 complex formation to deacetylate E2F1, which suppresses E2F1-mediated apoptotic gene expression in response to DNA damage[13]. KAP1 is also involved in MDM2-p53-HDAC1 complex and promotes deacetylation and MDM2-mediated degradation of p53[75]. More recently, melanoma antigen (MAGE) family proteins, which are highly expressed in many tumors, have been shown to enhance the formation of KAP1-p53 complex and to reduce p53 acetylation[80]. Interestingly, another study shows that MAGE proteins enhance the ubiquitin E3 ligase activity of KAP1 to ubiquitylate p53 for degradation[23]. These studies provide a rationale for developing compounds blocking KAP1-MAGE interaction for anti-cancer purpose[81].
In addition to regulating the activity of deacetylase complex, KAP1 is reported to disrupt the interaction of NF-κB and p300, an acetyltransferase. By this mechanism, the acetylation level of NF-κB is reduced and the transcriptional activity of NF-κB is dampened[51]. Besides, the physical interaction between KAP1 and STAT family members has been identified, and KAP1 negatively regulates STAT1 and STAT3 signaling[50,82]. However, the molecular mechanism underlying this event remains unclear. Whether KAP1 regulates the PTMs of STAT family still needs to be examined.
NON-TRANSCRIPTIONAL FUNCTIONS OF KAP1
While extensive efforts have been made in understanding how KAP1 regulates transcription, less is known about its non-transcriptional functions. In fact, KAP1 has a critical, transcription-independent role in DNA repair processes. Additionally, emerging evidence shows that KAP1 possesses enzymatic activity required for multiple cellular processes. In the following sections, we aim to discuss the role of KAP1 as a signaling scaffold protein in DNA repair and its novel roles as SUMO and ubiquitin E3 ligases.
Signaling scaffold protein in DNA damage response
White and colleagues identified that the PIKK family members, in response to DNA damage, phosphorylate KAP1 at serine-824. The serine-824 phosphorylated KAP1 co-localizes with several DNA repair factors, including γH2AX, 53BP1 and TopBP1, implicating a role for KAP1 in DNA repair processes[20,38]. It was further demonstrated that the KAP1 serine-824 phosphorylation is responsible for ATM-mediated chromatin relaxation, a crucial step for DNA double-strand break (DSB) repair[20]. Several lines of evidence suggest that the function of KAP1 in DSB repair is likely to be associated with the chromatin complexity and cell cycle status. Approximately 25% of DSBs that are located within heterochromatin require ATM signaling for repair, and knockdown of KAP1 bypasses the repair defects caused by ATM inhibition, suggesting that KAP1 is a direct downstream effector in ATM-mediated heterochromatin repair[83]. Although it has not been directly demonstrated, the SUMOylation of KAP1 seems to be important for heterochromatin maintenance because the interaction between KAP1 and the nucleosome remodeler, CHD3 requires KAP1 SUMOylation, and the chromatin retention of CHD3 is critical for chromatin plasticity during DDR[15,21,37]. It has been shown that the ATM-mediated KAP1 serine- 824 phosphorylation perturbs the SUMO-dependent interaction of KAP1 and CHD3 at the carboxyl-terminus of KAP1, thereby resulting in the de-condensation of heterochromatin[21]. Another possibility would be the deSUMOylase, SENP7, negatively regulates the SUMOylation status of KAP1 to release CHD3 and promote chromatin relaxation[37]. However, how SENP7 is recruited to deSUMOylate KAP1 at the damage sites remains to be defined. It is also suggested that following the KAP1 phosphorylation-dependent chromatin relaxation, the KAP1-dependent heterochromatin reconstitution mediated by the release of ATM is a prerequisite for error-free homologous recombination (HR) repair[84]. It would be interesting to further delineate how KAP1 is involved in the heterochromatin reconstitution and whether the re-SUMOylation of KAP1 is required for the reconstitution. In addition to its role in HR repair within heterochromatin, KAP1 has been shown to promote non-homologous end joining (NHEJ) repair, presumably within euchromatin[41]. Whether the chromatin localization of KAP1 determines its function in DSB repair is still unclear. Several studies have shown that HP1 is required for recruiting KAP1 to DNA damage sites for the repair within heterochromatin[44,85]. The disruption of HP1BD in KAP1 results in a defect in forming discrete serine-824 phosphorylated KAP1 foci, which have been considered as a critical signal for DSB repair[44]. How HP1 recruits KAP1 and whether KAP1 is responsible for HP1-mediated DSB repair warrant more detailed investigation.
Depletion of KAP1 is able to rescue the defects in NHEJ repair caused by ATM inhibition in G1 cells[83,86], whereas HR repair can be restored by knocking down KAP1 in ATM-inhibited G2 cells[87,88], indicating that the participation of KAP1 in specific DSB repair pathways is cell cycle-dependent. It is suggested that KAP1 serine-824 phosphorylation is enhanced by 53BP1 within heterochromatic regions and the concentrated phosphorylated KAP1 signal enables NHEJ repair[86]. On the other hand, a recent report shows that 53BP1 is required for enhancing KAP1 serine-824 phosphorylation and HR repair during G2-phase[89]. Collectively, these studies all indicate an important role of KAP1 in DSB repair. However, it is still unclear how cell cycle progression affects the role of KAP1 in selecting DSB repair pathway. Intriguingly, it has been observed that the retention of KAP1 on chromatin is largely reduced in G2-phase[87,90], suggesting that the association of KAP1 with chromatin is possibly regulated in a cell cycle-dependent manner and the chromatin retention of KAP1 may be a critical determinant that directs DSB pathway choice.
Taken together, chromatin remodeling could be one of the major functions served by KAP1 in DSB repair. Because KAP1 has multiple domains for protein-protein interaction, we speculate that KAP1 has additional roles, independent of chromatin remodeling, in DSB repair, such as the recruitment of members of DNA repair machinery.
Enzymatic activity
Recently, KAP1 has been found to possess SUMO E3 ligase activity via its PHD domain to recruit the SUMO-conjugating enzyme UBC9. This was first identified by demonstrating that KAP1 auto-SUMOylated its bromodomain to generate a repressive form of KAP1[15]. Later, another two proteins IRF7 and Vps34 have been identified as substrates of KAP1-mediated SUMOylation[24,25]. IRF7 is a transcription factor and master regulator of type I interferon-dependent immune responses. The SUMOylation of IRF7 by KAP1 RING finger domain reduces its transcription activity. Therefore, KAP1 could be a negative regulator of IRF7 and suppress IFN-based antiviral responses[24]. IRF7 is so far the only transcription factor identified as KAP1 target for SUMOylation. Several transcription factors are known to be SUMOylated and growing evidence suggests SUMOylation negatively regulates transcription[91]. Whether KAP1 mediates SUMOylation-dependent suppression of other transcription factors deserves another look.
More interestingly, KAP1 not only SUMOylates nuclear proteins but also targets a cytoplasmic protein, vacuolar protein-sorting(Vps) 34, which is crucial for autophagosome formation and plays a central role during autophagy. The SUMOylation of Vps34 enhances its binding to Beclin 1 and triggers autophagosome formation in the presence of acetylated HSP70. This study also suggests that the accumulation of KAP1 in the cytoplasm of cells treated with a pan-HDAC inhibitor, panobinostat via an unknown mechanism[25].
In addition to SUMO E3 ligase, the RING finger domain of KAP1 has ubiquitin E3 ligase activity. Studies have demonstrated that KAP1, in the presence of MAGE, ubiquitinates p53 and ZNF382 to facilitate their degradation[23,80,92]. This could be an additional mechanism for KAP1 to regulate gene expression. Taken together, recent progresses suggest a non-canonical function of KAP1, dependent on its SUMO and ubiquitin E3 ligase activity. These novel functions of KAP1 are not exclusively confined in the nucleus and might impact cellular physiology in response to various stresses by mechanisms beyond transcriptional regulation.
IMPLICATIONS OF KAP1 IN CELLULAR PHYSIOLOGY
KAP1 is involved in many aspects of cellular physiology (Figure 1). First, KAP1 exerts critical function during embryonic development because global Kap1-knockout causes embryonic lethality in mice due to the inability to undergo gastrulation[19] (Table 3). The function of KAP1 in maintaining pluripotency of embryonic stem cells (ESCs) has been demonstrated[78,93]. In addition, KAP1 is also required for ESC differentiation[94-97]. Studies using conditional Kap1-knockout mice show that KAP1 plays pivotal roles in different cellular maturation processes such as spermatogenesis, erythropoiesis, and the development of T-cell and B-cell (Table 3)[71,94,98-101]. Beyond regulating T-cell and B-cell differentiation, KAP1 also functions in immune response by additional mechanisms. For example, KAP1 is present in FOXP3-containing complex and facilitates the suppressor activity of regulatory T cells[74]. Moreover, KAP1 is involved in immunoglobulin class switch recombination[102]. Recent studies demonstrate that KAP1 associates with STAT1 and STAT3, master regulators of immune response, to negatively regulate their downstream signaling. These results suggest the involvement of KAP1 in immune response[50,51,82].
Table 3.
Mouse models illustrating the physiological functions of Kap1
| Animal model | Phenotype | Ref. |
| Kap1 knockout mice | Embryonic lethal prior to gastulation | [19] |
| Hemato-specific Kap1 knockout mice | Impaired erythropoiesis | [71,100] |
| T-cell-specific Kap1 knockout mice | Defective T-cell differentiation | [48,98] |
| B-cell-specific Kap1 knockout mice | Defective B-cell differentiation | [99] |
| Tamoxifen-inducible-germ cell-lineage-specific Kap1 depletion mice | Impaired spermatogenesis | [101] |
| Liver-specific Kap1 knockout mice | Male-predominant steatosis and hepatic adenoma | [120] |
| Kap1 knockout in mice forebrain | Anxiety-like-behavior and cognitive impairments | [103] |
Using a mouse model, Jakobsson et al[103] show that conditional deletion of Kap1 in adult forebrain caused higher level of anxiety-like activity, suggesting that Kap1 is a regulator for behavioral stress response. Although the molecular mechanism remains elusive, this could be due to the function of Kap1 in regulating gene expression in hippocampus. It is worth noting that among these dysregulated genes in hippocampus, some are imprinted genes[103]. We speculate that Kap1 is also required for the maintenance of a set of specific imprinted genes because many recent studies have demonstrated that KAP1 mediates DNA methylation by recruiting DNMTs to ICRs during early embryogenesis[59,61,62].
More than maintaining normal cellular physiological functions, KAP1 also regulates several pathways in response to different stresses. As mentioned earlier, several studies have focused on the role of KAP1 in DDR. KAP1 is known to be phosphorylated at serine-824 by ATM upon DNA damage and to transduce downstream signaling[20,38]. For instance, ATM-mediated KAP1 phosphorylation leads to de-repression of p21, Gadd45α, Bax, Puma, and Noxa, causing cell cycle arrest and apoptosis[16,17,36]. Additionally, KAP1 is an ATM downstream mediator during DSB-induced heterochromatin relaxation, which facilitates DNA repair[20,21,83]. KAP1 also cooperates with MDM2 to suppress p53 signaling by promoting p53 degradation[75,104]. Thus, PTMs of KAP1 might be important in regulating p53 activity as well.
In contrast to its co-repressor function, KAP1 can also be a co-activator in specific circumstances. It has been reported that mouse Kap1 interacts with Nrf2 to enhance Nrf2-meidated-cytoprotective function in NIH3T3 cells in response to oxidative stress[76]. However, whether KAP1 exerts similar co-activator activity in human cells is not clear.
Some studies demonstrate that KAP1 mediates viral gene expression and plays a role during viral latency. Specifically, KAP1 is associated with KSHV lytic gene promoter to suppress lytic gene expression and in turn maintains virus latency[57]. Similar observation has also been made in retroviruses such as Murine Leukemia Virus (MLV) and human T-cell lymphotropic virus-1 (HTLV-1), that KAP1 restricts pro-viral gene activation[105-108]. Interestingly, recent studies show that retroelements derived from endogenous retroviruses (ERVs), which are extensively present in mammalian genome, can also be silenced by KAP1 to protect genome integrity in embryonic stem cells[109-112]. In addition, KAP1-HDAC1 complex deacetylates and inhibits human immunodeficiency virus (HIV) integrase activity, thereby reducing HIV infectivity and its integration to host genome[113].
CLINICAL RELEVANCE FOCUSED ON CANCER BIOLOGY
The clinical relevance of KAP1 in diseases remains elusive. Most reports were based on studies in cancers. Higher expression of KAP1 has been linked to pro-metastatic cervical cancer[114]. Moreover, the up-regulation of KAP1 could be a potential marker in colorectal cancer patients and higher level of KAP1 is correlated with poorer overall survival in patients with gastric cancer and thyroid carcinoma[115-119]. These clinical studies suggest that higher KAP1 level is linked to a poor prognosis in certain cancers. However, opposite conclusions were also reported. KAP1 overexpression is associated with better overall survival in early-stage lung cancer[14]. Furthermore, a mouse model showed that Kap1-depletion in liver increases male-predominant hepatic adenoma[120] (Table 2).
Based on the studies focusing on transcriptional regulation, KAP1 suppresses p53 transcription activity and has been proposed to be a target for anti-cancer therapy[75,104]. Nonetheless, KAP1 also suppresses activity of oncogenic transcription factors HIF-1α and STAT3[72,82]. In vitro studies using cancer cell lines also show divergent results. KAP1 has been shown to restrain cell growth in breast and lung cancer cells whereas it promotes melanoma cell growth and enhanced KAP1 activity mediates cervical cancer invasion[14,80,114]. Taken together, it is still inconclusive how KAP1 regulates tumorigenesis. Because KAP1 is a multi-faceted protein, it might have tissue-specific function. More study is definitely required to understand how KAP1 contributes to tumorigenesis in particular tissue and whether KAP1 could be a target for cancer therapy.
CONCLUSION
The functions of KAP1 in diverse cellular physiology have been studied for 18 years. However, the mechanisms of KAP1 in regulating these different cellular processes are still largely unclear. The best-characterized role of KAP1 is its co-repressor function in regulating KRAB-ZFP-mediated or other transcription factor-associated gene silencing. In the future, the identification of more KAP1-interacting transcription factors may help to elucidate how KAP1 regulates gene expression under specific condition. Clearly, KAP1 is critical for maintaining genome integrity as KAP1 modulates the dynamics of hetero- and euchromatin maintenance. In addition, DNA damage induces a robust and transient KAP1 serine-824 phosphorylation and KAP1 is involved in DNA repair pathway choice. However, the molecular mechanism for KAP1-mediated DNA repair requires more rigorous investigation.
KAP1 is subjected to several types of PTM. The phosphorylation at serine-824 and SUMOylation at several lysine residues of KAP1 have been linked to gene regulation. Whether these lysine residues are subjected to additional PTMs and if yes, what is the associated functional consequences need to be further elucidated. Moreover, it would be interesting to look at other PTM sites, such as the phosphorylation of serine-473. Whether the serine-473 phosphorylation affects the transcriptional repressor activity of KAP1 is still under investigation. Another interesting question is how KAP1 shuttles among different cellular compartments by PTMs. The serine-824 phosphorylation has distinct nuclear localization pattern from the serine-473 phosphorylation, thus suggesting that KAP1 phosphorylation may affect its subcellular localization. Intriguingly, KAP1 is not only present in nucleus, and has been observed to translocate from nucleus to cytoplasm[25]. How PTM affects its nucleus-cytoplasmic shuttling and potential chromatin extraction of KAP1 deserve additional investigation. Finally, considering that KAP1 possesses SUMO and ubiquitin E3 activities, it might regulate the function or fate of its SUMO- and ubiquitin-targets. In the future, studying the non-transcriptional function of KAP1 might help to further decipher its unrecognized face.
Footnotes
P- Reviewer: Bartova E, Chen S, de la Serna IL, Zhu X S- Editor: Wen LL L- Editor: A E- Editor: Lu YJ
References
- 1.Le Douarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 3.Kim SS, Chen YM, O’Leary E, Witzgall R, Vidal M, Bonventre JV. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci USA. 1996;93:15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moosmann P, Georgiev O, Le Douarin B, Bourquin JP, Schaffner W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res. 1996;24:4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Krüppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Underhill C, Qutob MS, Yee SP, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem. 2000;275:40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- 8.Schultz DC, Friedman JR, Rauscher FJ. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satou A, Taira T, Iguchi-Ariga SM, Ariga H. A novel transrepression pathway of c-Myc. Recruitment of a transcriptional corepressor complex to c-Myc by MM-1, a c-Myc-binding protein. J Biol Chem. 2001;276:46562–46567. doi: 10.1074/jbc.M104937200. [DOI] [PubMed] [Google Scholar]
- 11.Satou A, Hagio Y, Taira T, Iguchi-Ariga SM, Ariga H. Repression of the c-fms gene in fibroblast cells by c-Myc-MM-1-TIF1beta complex. FEBS Lett. 2004;572:211–215. doi: 10.1016/j.febslet.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 12.Hagio Y, Kimura Y, Taira T, Fujioka Y, Iguchi-Ariga SM, Ariga H. Distinct localizations and repression activities of MM-1 isoforms toward c-Myc. J Cell Biochem. 2006;97:145–155. doi: 10.1002/jcb.20619. [DOI] [PubMed] [Google Scholar]
- 13.Wang C, Rauscher FJ, Cress WD, Chen J. Regulation of E2F1 function by the nuclear corepressor KAP1. J Biol Chem. 2007;282:29902–29909. doi: 10.1074/jbc.M704757200. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Chen DT, Kurtyka C, Rawal B, Fulp WJ, Haura EB, Cress WD. Tripartite motif containing 28 (Trim28) can regulate cell proliferation by bridging HDAC1/E2F interactions. J Biol Chem. 2012;287:40106–40118. doi: 10.1074/jbc.M112.380865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YK, Thomas SN, Yang AJ, Ann DK. Doxorubicin down-regulates Kruppel-associated box domain-associated protein 1 sumoylation that relieves its transcription repression on p21WAF1/CIP1 in breast cancer MCF-7 cells. J Biol Chem. 2007;282:1595–1606. doi: 10.1074/jbc.M606306200. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Lee YK, Jeng JC, Yen Y, Schultz DC, Shih HM, Ann DK. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem. 2007;282:36177–36189. doi: 10.1074/jbc.M706912200. [DOI] [PubMed] [Google Scholar]
- 18.Mascle XH, Germain-Desprez D, Huynh P, Estephan P, Aubry M. Sumoylation of the transcriptional intermediary factor 1beta (TIF1beta), the Co-repressor of the KRAB Multifinger proteins, is required for its transcriptional activity and is modulated by the KRAB domain. J Biol Chem. 2007;282:10190–10202. doi: 10.1074/jbc.M611429200. [DOI] [PubMed] [Google Scholar]
- 19.Cammas F, Mark M, Dollé P, Dierich A, Chambon P, Losson R. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development. 2000;127:2955–2963. doi: 10.1242/dev.127.13.2955. [DOI] [PubMed] [Google Scholar]
- 20.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 21.Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol. 2011;18:831–839. doi: 10.1038/nsmb.2077. [DOI] [PubMed] [Google Scholar]
- 22.Liu B, Wang Z, Ghosh S, Zhou Z. Defective ATM-Kap-1-mediated chromatin remodeling impairs DNA repair and accelerates senescence in progeria mouse model. Aging Cell. 2013;12:316–318. doi: 10.1111/acel.12035. [DOI] [PubMed] [Google Scholar]
- 23.Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang Q, Deng H, Li X, Wu X, Tang Q, Chang TH, Peng H, Rauscher FJ, Ozato K, Zhu F. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J Immunol. 2011;187:4754–4763. doi: 10.4049/jimmunol.1101704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y, Fiskus W, Yong B, Atadja P, Takahashi Y, Pandita TK, Wang HG, Bhalla KN. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc Natl Acad Sci USA. 2013;110:6841–6846. doi: 10.1073/pnas.1217692110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuda E, Agata Y, Sugai M, Katakai T, Gonda H, Shimizu A. Targeting of Krüppel-associated box-containing zinc finger proteins to centromeric heterochromatin. Implication for the gene silencing mechanisms. J Biol Chem. 2001;276:14222–14229. doi: 10.1074/jbc.M010663200. [DOI] [PubMed] [Google Scholar]
- 27.Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem. 2011;286:26267–26276. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng H, Gibson LC, Capili AD, Borden KL, Osborne MJ, Harper SL, Speicher DW, Zhao K, Marmorstein R, Rock TA, et al. The structurally disordered KRAB repression domain is incorporated into a protease resistant core upon binding to KAP-1-RBCC domain. J Mol Biol. 2007;370:269–289. doi: 10.1016/j.jmb.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 29.Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, Speicher DW, Rauscher FJ. Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J Mol Biol. 2000;295:1139–1162. doi: 10.1006/jmbi.1999.3402. [DOI] [PubMed] [Google Scholar]
- 31.Iyengar S, Ivanov AV, Jin VX, Rauscher FJ, Farnham PJ. Functional analysis of KAP1 genomic recruitment. Mol Cell Biol. 2011;31:1833–1847. doi: 10.1128/MCB.01331-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venturini L, You J, Stadler M, Galien R, Lallemand V, Koken MH, Mattei MG, Ganser A, Chambon P, Losson R, et al. TIF1gamma, a novel member of the transcriptional intermediary factor 1 family. Oncogene. 1999;18:1209–1217. doi: 10.1038/sj.onc.1202655. [DOI] [PubMed] [Google Scholar]
- 33.Lechner MS, Begg GE, Speicher DW, Rauscher FJ. Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol Cell Biol. 2000;20:6449–6465. doi: 10.1128/mcb.20.17.6449-6465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riclet R, Chendeb M, Vonesch JL, Koczan D, Thiesen HJ, Losson R, Cammas F. Disruption of the interaction between transcriptional intermediary factor 1{beta} and heterochromatin protein 1 leads to a switch from DNA hyper- to hypomethylation and H3K9 to H3K27 trimethylation on the MEST promoter correlating with gene reactivation. Mol Biol Cell. 2009;20:296–305. doi: 10.1091/mbc.E08-05-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Lin HH, Chen H, Xu X, Shih HM, Ann DK. SUMOylation of the transcriptional co-repressor KAP1 is regulated by the serine and threonine phosphatase PP1. Sci Signal. 2010;3:ra32. doi: 10.1126/scisignal.2000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garvin AJ, Densham RM, Blair-Reid SA, Pratt KM, Stone HR, Weekes D, Lawrence KJ, Morris JR. The deSUMOylase SENP7 promotes chromatin relaxation for homologous recombination DNA repair. EMBO Rep. 2013;14:975–983. doi: 10.1038/embor.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White DE, Negorev D, Peng H, Ivanov AV, Maul GG, Rauscher FJ. KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res. 2006;66:11594–11599. doi: 10.1158/0008-5472.CAN-06-4138. [DOI] [PubMed] [Google Scholar]
- 39.Tomimatsu N, Mukherjee B, Burma S. Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep. 2009;10:629–635. doi: 10.1038/embor.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee DH, Goodarzi AA, Adelmant GO, Pan Y, Jeggo PA, Marto JA, Chowdhury D. Phosphoproteomic analysis reveals that PP4 dephosphorylates KAP-1 impacting the DNA damage response. EMBO J. 2012;31:2403–2415. doi: 10.1038/emboj.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Xu L, Zhong J, Liao J, Li J, Xu X. Protein phosphatase PP4 is involved in NHEJ-mediated repair of DNA double-strand breaks. Cell Cycle. 2012;11:2643–2649. doi: 10.4161/cc.20957. [DOI] [PubMed] [Google Scholar]
- 42.Hu C, Zhang S, Gao X, Gao X, Xu X, Lv Y, Zhang Y, Zhu Z, Zhang C, Li Q, et al. Roles of Kruppel-associated Box (KRAB)-associated Co-repressor KAP1 Ser-473 Phosphorylation in DNA Damage Response. J Biol Chem. 2012;287:18937–18952. doi: 10.1074/jbc.M111.313262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolderson E, Savage KI, Mahen R, Pisupati V, Graham ME, Richard DJ, Robinson PJ, Venkitaraman AR, Khanna KK. Kruppel-associated Box (KRAB)-associated co-repressor (KAP-1) Ser-473 phosphorylation regulates heterochromatin protein 1β (HP1-β) mobilization and DNA repair in heterochromatin. J Biol Chem. 2012;287:28122–28131. doi: 10.1074/jbc.M112.368381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White D, Rafalska-Metcalf IU, Ivanov AV, Corsinotti A, Peng H, Lee SC, Trono D, Janicki SM, Rauscher FJ. The ATM substrate KAP1 controls DNA repair in heterochromatin: regulation by HP1 proteins and serine 473/824 phosphorylation. Mol Cancer Res. 2012;10:401–414. doi: 10.1158/1541-7786.MCR-11-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blasius M, Forment JV, Thakkar N, Wagner SA, Choudhary C, Jackson SP. A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 2011;12:R78. doi: 10.1186/gb-2011-12-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang CW, Chou HY, Lin YS, Huang KH, Chang CJ, Hsu TC, Lee SC. Phosphorylation at Ser473 regulates heterochromatin protein 1 binding and corepressor function of TIF1beta/KAP1. BMC Mol Biol. 2008;9:61. doi: 10.1186/1471-2199-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou XF, Yu J, Chang M, Zhang M, Zhou D, Cammas F, Sun SC. TRIM28 mediates chromatin modifications at the TCRα enhancer and regulates the development of T and natural killer T cells. Proc Natl Acad Sci USA. 2012;109:20083–20088. doi: 10.1073/pnas.1214704109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chikuma S, Suita N, Okazaki IM, Shibayama S, Honjo T. TRIM28 prevents autoinflammatory T cell development in vivo. Nat Immunol. 2012;13:596–603. doi: 10.1038/ni.2293. [DOI] [PubMed] [Google Scholar]
- 49.King CA. Kaposi’s sarcoma-associated herpesvirus kaposin B induces unique monophosphorylation of STAT3 at serine 727 and MK2-mediated inactivation of the STAT3 transcriptional repressor TRIM28. J Virol. 2013;87:8779–8791. doi: 10.1128/JVI.02976-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamitani S, Ohbayashi N, Ikeda O, Togi S, Muromoto R, Sekine Y, Ohta K, Ishiyama H, Matsuda T. KAP1 regulates type I interferon/STAT1-mediated IRF-1 gene expression. Biochem Biophys Res Commun. 2008;370:366–370. doi: 10.1016/j.bbrc.2008.03.104. [DOI] [PubMed] [Google Scholar]
- 51.Kamitani S, Togi S, Ikeda O, Nakasuji M, Sakauchi A, Sekine Y, Muromoto R, Oritani K, Matsuda T. Krüppel-associated box-associated protein 1 negatively regulates TNF-α-induced NF-κB transcriptional activity by influencing the interactions among STAT3, p300, and NF-κB/p65. J Immunol. 2011;187:2476–2483. doi: 10.4049/jimmunol.1003243. [DOI] [PubMed] [Google Scholar]
- 52.Eames HL, Saliba DG, Krausgruber T, Lanfrancotti A, Ryzhakov G, Udalova IA. KAP1/TRIM28: an inhibitor of IRF5 function in inflammatory macrophages. Immunobiology. 2012;217:1315–1324. doi: 10.1016/j.imbio.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 53.Kubota S, Fukumoto Y, Aoyama K, Ishibashi K, Yuki R, Morinaga T, Honda T, Yamaguchi N, Kuga T, Tomonaga T, et al. Phosphorylation of KRAB-associated protein 1 (KAP1) at Tyr-449, Tyr-458, and Tyr-517 by nuclear tyrosine kinases inhibits the association of KAP1 and heterochromatin protein 1α (HP1α) with heterochromatin. J Biol Chem. 2013;288:17871–17883. doi: 10.1074/jbc.M112.437756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai IL, Lin TP, Yao YL, Lin CY, Hsieh MJ, Yang WM. Histone deacetylase 10 relieves repression on the melanogenic program by maintaining the deacetylation status of repressors. J Biol Chem. 2010;285:7187–7196. doi: 10.1074/jbc.M109.061861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Geen H, Squazzo SL, Iyengar S, Blahnik K, Rinn JL, Chang HY, Green R, Farnham PJ. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 2007;3:e89. doi: 10.1371/journal.pgen.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Dénervaud N, Bucher P, Trono D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang PC, Fitzgerald LD, Van Geelen A, Izumiya Y, Ellison TJ, Wang DH, Ann DK, Luciw PA, Kung HJ. Kruppel-associated box domain-associated protein-1 as a latency regulator for Kaposi’s sarcoma-associated herpesvirus and its modulation by the viral protein kinase. Cancer Res. 2009;69:5681–5689. doi: 10.1158/0008-5472.CAN-08-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowbotham SP, Barki L, Neves-Costa A, Santos F, Dean W, Hawkes N, Choudhary P, Will WR, Webster J, Oxley D, et al. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol Cell. 2011;42:285–296. doi: 10.1016/j.molcel.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 59.Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Rep. 2012;2:766–773. doi: 10.1016/j.celrep.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuo X, Sheng J, Lau HT, McDonald CM, Andrade M, Cullen DE, Bell FT, Iacovino M, Kyba M, Xu G, et al. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J Biol Chem. 2012;287:2107–2118. doi: 10.1074/jbc.M111.322644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takikawa S, Wang X, Ray C, Vakulenko M, Bell FT, Li X. Human and mouse ZFP57 proteins are functionally interchangeable in maintaining genomic imprinting at multiple imprinted regions in mouse ES cells. Epigenetics. 2013;8:1268–1279. doi: 10.4161/epi.26544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- 64.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 65.Kacem S, Feil R. Chromatin mechanisms in genomic imprinting. Mamm Genome. 2009;20:544–556. doi: 10.1007/s00335-009-9223-4. [DOI] [PubMed] [Google Scholar]
- 66.Itokawa Y, Yanagawa T, Yamakawa H, Watanabe N, Koga H, Nagase T. KAP1-independent transcriptional repression of SCAN-KRAB-containing zinc finger proteins. Biochem Biophys Res Commun. 2009;388:689–694. doi: 10.1016/j.bbrc.2009.08.065. [DOI] [PubMed] [Google Scholar]
- 67.Meylan S, Groner AC, Ambrosini G, Malani N, Quenneville S, Zangger N, Kapopoulou A, Kauzlaric A, Rougemont J, Ciuffi A, et al. A gene-rich, transcriptionally active environment and the pre-deposition of repressive marks are predictive of susceptibility to KRAB/KAP1-mediated silencing. BMC Genomics. 2011;12:378. doi: 10.1186/1471-2164-12-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jin VX, O’Geen H, Iyengar S, Green R, Farnham PJ. Identification of an OCT4 and SRY regulatory module using integrated computational and experimental genomics approaches. Genome Res. 2007;17:807–817. doi: 10.1101/gr.6006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng L, Pan H, Li S, Flesken-Nikitin A, Chen PL, Boyer TG, Lee WH. Sequence-specific transcriptional corepressor function for BRCA1 through a novel zinc finger protein, ZBRK1. Mol Cell. 2000;6:757–768. doi: 10.1016/s1097-2765(00)00075-7. [DOI] [PubMed] [Google Scholar]
- 70.Takahashi K, Sugi Y, Hosono A, and Kaminogawa S. Epigenetic Regulation of TLR4 Gene Expression in Intestinal Epithelial Cells for the Maintenance of Intestinal Homeostasis. J Immunol. 2009;183:6522–6529. doi: 10.4049/jimmunol.0901271. [DOI] [PubMed] [Google Scholar]
- 71.Barde I, Rauwel B, Marin-Florez RM, Corsinotti A, Laurenti E, Verp S, Offner S, Marquis J, Kapopoulou A, Vanicek J, et al. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science. 2013;340:350–353. doi: 10.1126/science.1232398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Z, Wang D, Na X, Schoen SR, Messing EM, Wu G. The VHL protein recruits a novel KRAB-A domain protein to repress HIF-1alpha transcriptional activity. EMBO J. 2003;22:1857–1867. doi: 10.1093/emboj/cdg173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng H, Ivanov AV, Oh HJ, Lau YF, Rauscher FJ. Epigenetic gene silencing by the SRY protein is mediated by a KRAB-O protein that recruits the KAP1 co-repressor machinery. J Biol Chem. 2009;284:35670–35680. doi: 10.1074/jbc.M109.032086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang C, Martin S, Pfleger C, Du J, Buckner JH, Bluestone JA, Riley JL, Ziegler SF. Cutting Edge: a novel, human-specific interacting protein couples FOXP3 to a chromatin-remodeling complex that contains KAP1/TRIM28. J Immunol. 2013;190:4470–4473. doi: 10.4049/jimmunol.1203561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang C, Ivanov A, Chen L, Fredericks WJ, Seto E, Rauscher FJ, Chen J. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J. 2005;24:3279–3290. doi: 10.1038/sj.emboj.7600791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maruyama A, Nishikawa K, Kawatani Y, Mimura J, Hosoya T, Harada N, Yamamato M, Itoh K. The novel Nrf2-interacting factor KAP1 regulates susceptibility to oxidative stress by promoting the Nrf2-mediated cytoprotective response. Biochem J. 2011;436:387–397. doi: 10.1042/BJ20101748. [DOI] [PubMed] [Google Scholar]
- 77.Tian C, Xing G, Xie P, Lu K, Nie J, Wang J, Li L, Gao M, Zhang L, He F. KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol. 2009;11:580–591. doi: 10.1038/ncb1864. [DOI] [PubMed] [Google Scholar]
- 78.Seki Y, Kurisaki A, Watanabe-Susaki K, Nakajima Y, Nakanishi M, Arai Y, Shiota K, Sugino H, Asashima M. TIF1beta regulates the pluripotency of embryonic stem cells in a phosphorylation-dependent manner. Proc Natl Acad Sci USA. 2010;107:10926–10931. doi: 10.1073/pnas.0907601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takahashi K, Sugi Y, Hosono A, Kaminogawa S. Epigenetic regulation of TLR4 gene expression in intestinal epithelial cells for the maintenance of intestinal homeostasis. J Immunol. 2009;183:6522–6529. doi: 10.4049/jimmunol.0901271. [DOI] [PubMed] [Google Scholar]
- 80.Yang B, O’Herrin SM, Wu J, Reagan-Shaw S, Ma Y, Bhat KM, Gravekamp C, Setaluri V, Peters N, Hoffmann FM, et al. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer Res. 2007;67:9954–9962. doi: 10.1158/0008-5472.CAN-07-1478. [DOI] [PubMed] [Google Scholar]
- 81.Bhatia N, Yang B, Xiao TZ, Peters N, Hoffmann MF, Longley BJ. Identification of novel small molecules that inhibit protein-protein interactions between MAGE and KAP-1. Arch Biochem Biophys. 2011;508:217–221. doi: 10.1016/j.abb.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsuruma R, Ohbayashi N, Kamitani S, Ikeda O, Sato N, Muromoto R, Sekine Y, Oritani K, Matsuda T. Physical and functional interactions between STAT3 and KAP1. Oncogene. 2008;27:3054–3059. doi: 10.1038/sj.onc.1210952. [DOI] [PubMed] [Google Scholar]
- 83.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 84.Geuting V, Reul C, Löbrich M. ATM release at resected double-strand breaks provides heterochromatin reconstitution to facilitate homologous recombination. PLoS Genet. 2013;9:e1003667. doi: 10.1371/journal.pgen.1003667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baldeyron C, Soria G, Roche D, Cook AJ, Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol. 2011;193:81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Noon AT, Shibata A, Rief N, Löbrich M, Stewart GS, Jeggo PA, Goodarzi AA. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 87.Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Löbrich M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009;28:3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, Kakarougkas A, Meek K, Taucher-Scholz G, Löbrich M, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011;30:1079–1092. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kakarougkas A, Ismail A, Klement K, Goodarzi AA, Conrad S, Freire R, Shibata A, Lobrich M, Jeggo PA. Opposing roles for 53BP1 during homologous recombination. Nucleic Acids Res. 2013;41:9719–9731. doi: 10.1093/nar/gkt729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goodarzi AA, Noon AT, Jeggo PA. The impact of heterochromatin on DSB repair. Biochem Soc Trans. 2009;37:569–576. doi: 10.1042/BST0370569. [DOI] [PubMed] [Google Scholar]
- 91.Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiao TZ, Bhatia N, Urrutia R, Lomberk GA, Simpson A, Longley BJ. MAGE I transcription factors regulate KAP1 and KRAB domain zinc finger transcription factor mediated gene repression. PLoS One. 2011;6:e23747. doi: 10.1371/journal.pone.0023747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cammas F, Oulad-Abdelghani M, Vonesch JL, Huss-Garcia Y, Chambon P, Losson R. Cell differentiation induces TIF1beta association with centromeric heterochromatin via an HP1 interaction. J Cell Sci. 2002;115:3439–3448. doi: 10.1242/jcs.115.17.3439. [DOI] [PubMed] [Google Scholar]
- 95.Herzog M, Wendling O, Guillou F, Chambon P, Mark M, Losson R, Cammas F. TIF1β association with HP1 is essential for post-gastrulation development, but not for Sertoli cell functions during spermatogenesis. Dev Biol. 2011;350:548–558. doi: 10.1016/j.ydbio.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 96.Cammas F, Herzog M, Lerouge T, Chambon P, Losson R. Association of the transcriptional corepressor TIF1beta with heterochromatin protein 1 (HP1): an essential role for progression through differentiation. Genes Dev. 2004;18:2147–2160. doi: 10.1101/gad.302904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cammas F, Janoshazi A, Lerouge T, and Losson R. Dynamic and selective interactions of the transcriptional corepressor TIF1 beta with the heterochromatin protein HP1 isotypes during cell differentiation. Differentiation. 2007;75:627–637. doi: 10.1111/j.1432-0436.2007.00166.x. [DOI] [PubMed] [Google Scholar]
- 98.Santoni de Sio FR, Barde I, Offner S, Kapopoulou A, Corsinotti A, Bojkowska K, Genolet R, Thomas JH, Luescher IF, Pinschewer D, et al. KAP1 regulates gene networks controlling T-cell development and responsiveness. FASEB J. 2012;26:4561–4575. doi: 10.1096/fj.12-206177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Santoni de Sio FR, Massacand J, Barde I, Offner S, Corsinotti A, Kapopoulou A, Bojkowska K, Dagklis A, Fernandez M, Ghia P, et al. KAP1 regulates gene networks controlling mouse B-lymphoid cell differentiation and function. Blood. 2012;119:4675–4685. doi: 10.1182/blood-2011-12-401117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hosoya T, Clifford M, Losson R, Tanabe O, Engel JD. TRIM28 is essential for erythroblast differentiation in the mouse. Blood. 2013;122:3798–3807. doi: 10.1182/blood-2013-04-496166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weber P, Cammas F, Gerard C, Metzger D, Chambon P, Losson R, Mark M. Germ cell expression of the transcriptional co-repressor TIF1beta is required for the maintenance of spermatogenesis in the mouse. Development. 2002;129:2329–2337. doi: 10.1242/dev.129.10.2329. [DOI] [PubMed] [Google Scholar]
- 102.Jeevan-Raj BP, Robert I, Heyer V, Page A, Wang JH, Cammas F, Alt FW, Losson R, Reina-San-Martin B. Epigenetic tethering of AID to the donor switch region during immunoglobulin class switch recombination. J Exp Med. 2011;208:1649–1660. doi: 10.1084/jem.20110118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jakobsson J, Cordero MI, Bisaz R, Groner AC, Busskamp V, Bensadoun JC, Cammas F, Losson R, Mansuy IM, Sandi C, et al. KAP1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron. 2008;60:818–831. doi: 10.1016/j.neuron.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 104.Okamoto K, Kitabayashi I, Taya Y. KAP1 dictates p53 response induced by chemotherapeutic agents via Mdm2 interaction. Biochem Biophys Res Commun. 2006;351:216–222. doi: 10.1016/j.bbrc.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 105.Wolf D, Cammas F, Losson R, Goff SP. Primer binding site-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J Virol. 2008;82:4675–4679. doi: 10.1128/JVI.02445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 107.Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wolf D, Hug K, Goff SP. TRIM28 mediates primer binding site-targeted silencing of Lys1,2 tRNA-utilizing retroviruses in embryonic cells. Proc Natl Acad Sci USA. 2008;105:12521–12526. doi: 10.1073/pnas.0805540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 110.Schlesinger S, Lee AH, Wang GZ, Green L, Goff SP. Proviral silencing in embryonic cells is regulated by Yin Yang 1. Cell Rep. 2013;4:50–58. doi: 10.1016/j.celrep.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 112.Tan X, Xu X, Elkenani M, Smorag L, Zechner U, Nolte J, Engel W, Pantakani DV. Zfp819, a novel KRAB-zinc finger protein, interacts with KAP1 and functions in genomic integrity maintenance of mouse embryonic stem cells. Stem Cell Res. 2013;11:1045–1059. doi: 10.1016/j.scr.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 113.Allouch A, Di Primio C, Alpi E, Lusic M, Arosio D, Giacca M, Cereseto A. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe. 2011;9:484–495. doi: 10.1016/j.chom.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 114.Lin LF, Li CF, Wang WJ, Yang WM, Wang DD, Chang WC, Lee WH, Wang JM. Loss of ZBRK1 contributes to the increase of KAP1 and promotes KAP1-mediated metastasis and invasion in cervical cancer. PLoS One. 2013;8:e73033. doi: 10.1371/journal.pone.0073033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yokoe T, Toiyama Y, Okugawa Y, Tanaka K, Ohi M, Inoue Y, Mohri Y, Miki C, and Kusunoki M. KAP1 Is Associated With Peritoneal Carcinomatosis in Gastric Cancer. Ann Surg Oncol. 2009;17:821–828. doi: 10.1245/s10434-009-0795-8. [DOI] [PubMed] [Google Scholar]
- 116.Hector S, Chen H, Kijanka G, Murray F, Prehn JH. A reverse-ELISA for the detection of TRIM28/KAP1 serum autoantibodies in colorectal cancer patients. Acta Oncol. 2012;51:394–396. doi: 10.3109/0284186X.2011.652742. [DOI] [PubMed] [Google Scholar]
- 117.Wang YY, Li L, Zhao ZS, Wang HJ. Clinical utility of measuring expression levels of KAP1, TIMP1 and STC2 in peripheral blood of patients with gastric cancer. World J Surg Oncol. 2013;11:81. doi: 10.1186/1477-7819-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martins MB, Marcello MA, Morari EC, Cunha LL, Soares FA, Vassallo J, Ward LS. Clinical utility of KAP-1 expression in thyroid lesions. Endocr Pathol. 2013;24:77–82. doi: 10.1007/s12022-013-9245-z. [DOI] [PubMed] [Google Scholar]
- 119.Kijanka G, Hector S, Kay EW, Murray F, Cummins R, Murphy D, MacCraith BD, Prehn JH, Kenny D. Human IgG antibody profiles differentiate between symptomatic patients with and without colorectal cancer. Gut. 2010;59:69–78. doi: 10.1136/gut.2009.178574. [DOI] [PubMed] [Google Scholar]
- 120.Bojkowska K, Aloisio F, Cassano M, Kapopoulou A, Santoni de Sio F, Zangger N, Offner S, Cartoni C, Thomas C, Quenneville S, et al. Liver-specific ablation of Krüppel-associated box-associated protein 1 in mice leads to male-predominant hepatosteatosis and development of liver adenoma. Hepatology. 2012;56:1279–1290. doi: 10.1002/hep.25767. [DOI] [PMC free article] [PubMed] [Google Scholar]


