“a disaster of biblical proportions….Dogs and cats living together! Mass hysteria!”
Bill Murray as Dr. Peter Venkman in the 1984 film Ghostbusters, co-written by Harold Ramis and Dan Aykroyd.
The idea that cross-species co-habitation is catastrophic does not seem to apply to RNA. Post-transcriptional regulation of protein coding mRNA transcripts bound by ~20 nucleotide (nt) single stranded microRNAs is thoroughly established 1. Another species of non-coding RNA, designated long noncoding (lnc) RNAs because of their longer size (>200 nt), can regulate microRNA abundance by binding and sequestering them, acting as so-called “microRNA sponges” 2. In this issue of Circulation Research, Wang et al close the circle by describing a lncRNA-microRNA-mRNA trio that functions interdependently to regulate cardiac hypertrophy 3. The mRNA encodes the immune response adaptor protein Myd88 (Myeloid differentiation primary response 88), which regulates cardiomyocyte and cardiac hypertrophy via mechanisms that are not defined. Myd88 can be targetted by miR-489, steady state levels of which are decreased during angiotensin II-stimulated cardiomyocyte hypertrophy (thus de-repressing Myd88). And miR-489 can also be bound by lncRNA AK048451 (here named CHRF for Cardiac Hypertrophy Related Factor), levels of which are increased by angiotensin II; by sequestering miR-489 CHRF impairs the microRNA’s ability to downregulate Myd88 mRNA.
One of the revelations of multi-species genome sequencing is that organism complexity relates to the richness of the genetic regulatory machinery, not the number of protein coding genes. Much of the noncoding mammalian genome previously referred to as “junk DNA” is now recognized as regulatory. For example, we recognize that conventional DNA-binding trans-activating proteins 4 work in concert with microRNAs to orchestrate development of embryonic hearts and/or the response to cardiac stress 1.
MicroRNAs are one species of noncoding RNAs that include long non-coding (lnc) RNAs. Because RNAs do not possess intrinsic catalytic activity, biological function is directly or indirectly determined by nucleotide sequence. For example, ~20 nt linear microRNAs embedded with Argonaute proteins in RNA-induced silencing complexes (RISCs) bind via seed-sequence interactions to complementary nucleotides in 3′ untranslated regions (UTR) of protein coding mRNAs 5, 6, thereby recruiting them to the RISC for silencing and degradation. Primary microRNA nucleotide sequence is therefore a key determinant of mRNA targeting.
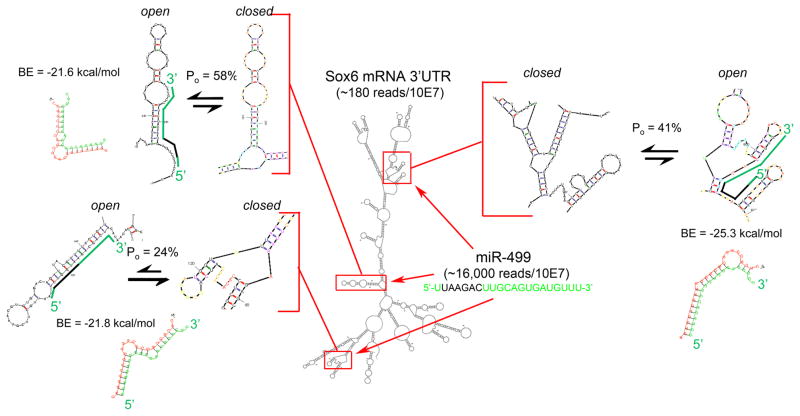
Most bioinformatic miR-mRNA sequence comparison algorithms like TargetScan (www.targetscan.org) do not consider the physical configurations resulting from pairing of internal complementary mRNA sequences. Obviously, in addition to having complementary sequence, a microRNA binding domain must be structured such that the critical seed sequence binding site is accessible, i.e. at least partially single-stranded. This is depicted in Figure 1 for miR-499 and its validated mRNA target, the transcription factor Sox67, 8. TargetScan6 identifies 4 potential miR-499 binding sites within the mouse Sox6 3′UTR, one of which is poorly conserved and thermodynamically unfavorable, and therefore not depicted. Structural modeling of the Sox6 3′UTR using Mfold (mfold.rit.albany.edu) reveals that ‘seed’-region binding sites of two of the three remaining binding domains are more accessible (i.e. have a greater open probability; Po = 58% and 41%) than the other (Po = 24%; Figure 1). mRNA structure is therefore another important determinant of miR-mRNA binding.
Figure 1. Schematic diagram of miR-499 binding to Sox6 mRNA.
An M-fold structure of Sox6 3′UTR is shown with miR-499 binding sites framed in red. These binding domains are enlarged as insets and shown in configurations less favorable (closed) and more favorable (open) for miR-499 seed sequence binding; probability of the more open structure is reported for each as Po. miR-499 is depicted on the open Sox6 configuration in green, with seed sequence in black; RNAhybrid duplex structure and minimum hybridization (binding) energy (BE) is given for each interaction. RNA mass/abundance values determined from publically available adult mouse heart RNA sequencing data are shown as reads/10 million reads.
Any RNA-RNA pairing event must follow the law of mass action, driven by the amounts of free microRNA and mRNA, and the RNA-RNA binding energy (analogous to binding affinity; lower binding energy reflects greater binding affinity) 9, 10. Thus, quantity of microRNA and mRNA, and the thermodynamic characteristics of their pairing are also key determinants of miR-mRNA binding. RNA mass is quantifiable by deep sequencing 11–13, and raw and aligned microRNA and mRNA mouse heart sequence data are publically available for interrogation (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE55792). This resource reports miR-499 levels as ~1600 RpM (Reads per Million miR reads) and Sox6 mRNA levels as ~3-6 FPKM (Fragments/reads Per Kb of exon per Million mRNA reads), an expression level typical for transcription factors. The minimum free energy of hybridization for miR-499 to the Sox6 3′UTR (calculated using RNAhybrid ([http://bibiserv.techfak.uni-bielefeld.de/rnahybrid]14) is less than −21 kcal/mol for each site, reflecting strong binding (Figure 1). Thus, miR-499 and Sox6 are both present at meaningful levels in mouse hearts, the structure of the mouse Sox6 3′UTR provides a high probability of access to at least two miR-499 binding sites, and miR-mRNA binding energy at these sites is compatible with formation of an RNA pair with sufficient stability to retain the mRNA at the microRNA-loaded RISC. Accordingly, Sox6 is highly (~5-fold) enriched in cardiac RISC complexes 13, 15.
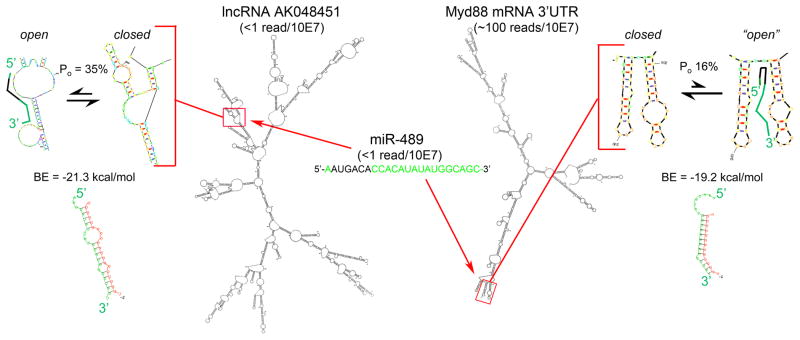
The same analytical tools can be applied to the lncRNA-microRNA-mRNA interaction described by Wang et al 3. TargetScan identifies Myd88 binding sites for at least 80 microRNAs, but not for miR-489 (likely because of very poor seed sequence complementarity). RNAhybrid analysis of the miR-489 site described by Wang reveals a healthy minimum free energy of hybridization between miR-489 and Myd88 of −19.2 kcal/mol. However, structural modeling of the Myd88 mRNA 3′UTR shows that the reported miR-489 binding site is relatively inaccessible (Po = only 16%), and that the “open” seed binding configuration is still relatively closed (Figure 2).
Figure 2. Schematic diagram of miR-489 binding to lncRNAAK048451 and Myd88 mRNA.
M-fold and RNAhybrid structures, Po and BE values, and abundance for the interacting RNA species reported by Wang et al, are depicted as in Figure 1.
High mass can compensate for low binding affinity; an improbable binding event can have physiological significance if the two binding partners are present at large quantities. According to the same web resource used above, Myd88 mRNA is expressed at levels similar to Sox6 mRNA (Figure 2). However, mouse heart miR-489 levels were not measurable (i.e. <1 read/10 million miR reads) in published reports 11, 12, and the miR-489 host gene (Calcr) is also present at <1 read/10 million. Thus, miR-489 is uncommon in normal mouse hearts. Given the unfavorable mRNA 3′UTR structure, its interaction with native Myd88 may also be uncommon.
We applied the same tools and resources to the miR-489 lncRNA AK048451 interaction (Figure 2). AK048451 is present in (poly-A selected) normal adult mouse heart mRNA at less than 1 read/10 million mRNA reads (although we consistently detect it at levels of 2-3 reads/10 million mapped reads in embryonic mouse hearts; unpublished data). Accessibility of the seed sequence binding site at the reported miR-489 binding domain in lncRNA AK048451 is comparable to the functional miR-499 sites in Sox6 (Po = 35%), with an equivalent minimum free binding energy (-21.3 kcal/mol). When this lncRNA is upregulated, as in angiotensin II-stimulated hypertrophy, it is likely to bind miR-489.
Limitations and assumptions of these analytical tools are significant: we assume that the measured level of each RNA species represents its “free” concentration in the cell of phenotypic interest (cardiomyocyte), and that the microRNA, mRNA, and lncRNA are not subject to subcellular compartmentalization, i.e. are mutually available to interact. The model also does not account for “sequestration binding” of miR-489 by any other lncRNAs or more abundant cardiac-expressed mRNAs with roughly equivalent binding energies for miR-489 (e.g. Mef2c: FPKM ~25, BE for miR-489 = −17.1 kcal/mol).
These analyses illustrate how important variables can be overlooked when an inherently rich and necessarily complex biologically regulatory mechanism is reduced to a linear concept. Not only will multiple variables not be unaccounted for, but each of the variables is context-dependent, transient, and probabilistic: 1. RNA-RNA interactions, whether intra-molecular binding that produces folded RNA structures or inter-molecular binding between microRNAs and their targets or “sponges”, are dynamic. Accordingly, RNA structures are ranked nondeterministically using either the “open probability” metric or by calculating the RNA-RNA minimum free energy of binding. 2. The steady-state levels of RNA species, whether coding and noncoding, are constantly changing because of modulated expression, processing, and sequestration. Absolute RNA content (mass), not relative RNA expression, is a key factor. These quantitative data are not provided by arrays or RT-qPCR. 3. Individual noncoding RNAs have multiple different (DNA and RNA) binding partners, and therefore exert more than one function. Indeed, the complex folded structures of lncRNAs produces striking mechanistic diversity, enabling them to function as protein anchors, as chaperones that target transcriptional modulators to specific genes, and as binding partners for other RNA species.
As shown above, web-based resources can predict RNA structure and thermodynamic parameters of RNA-RNA binding, and mRNA and small RNA sequencing data are increasingly obtainable. The NCBI GEO resource to which we uploaded results of over 40 individual normal adult mouse heart RNA and small RNA sequencing studies provides access to pre-processed data (facilitating interrogation of specific transcripts of interest) and ‘raw’ sequencing reads (for application of improved or alternate alignment algorithms).
We will inevitably uncover additional mechanisms that modify gene expression and transcript stability/function. A lncRNA acting as a microRNA sponge is the current example of one epigenetic mechanism regulating another, so-called “inter-epigenetic” regulation. Indeed, by directing proteins to specific genomic DNA sites, lncRNAs are well positioned to modify other epigenetic events, such as chromatin structure and DNA methylation. We propose that regulatory complexity be embraced: The observation that a given molecular or functional interaction can occur can be supported by analyses of the probability that it takes place in the relevant biological context. Unbiased genome-wide experimentation that quantitatively evaluates levels of, and interactions between, microRNAs, lncRNAs, and mRNAs can be a first step toward this goal. Agnostic results ranked according to probability estimates factoring in RNA structure, binding affinity, and RNA species abundance can be validated using standard co-precipitation and function reporter assays with binding site mutagenesis. Such an approach has the same advantages as did eschewing “candidate gene” analysis in favor of unbiased genome-wide discovery of disease-causing human DNA variants 16.
Acknowledgments
Supported by NIH R01 HL108943.
Footnotes
The authors declare no conflicts of interest.
References
- 1.Condorelli G, Latronico MV, Dorn GW., 2nd MicroRNAs in heart disease: Putative novel therapeutic targets? Eur Heart J. 2010;31:649–658. doi: 10.1093/eurheartj/ehp573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 3.Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, Liu CY, Sun T, Zhang XJ, Li PF. A long noncoding RNA, CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res. 2014 doi: 10.1161/CIRCRESAHA.114.302476. (In Press) [DOI] [PubMed] [Google Scholar]
- 4.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khorshid M, Hausser J, Zavolan M, van Nimwegen E. A biophysical miRNA-mRNA interaction model infers canonical and noncanonical targets. Nat Methods. 2013;10:253–255. doi: 10.1038/nmeth.2341. [DOI] [PubMed] [Google Scholar]
- 6.Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorn GW, 2nd, Matkovich SJ, Eschenbacher WH, Zhang Y. A human 3′ miR-499 mutation alters cardiac mRNA targeting and function. Circ Res. 2012;110:958–967. doi: 10.1161/CIRCRESAHA.111.260752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorn GW., 2nd Decoding the cardiac message: The 2011 Thomas W. Smith Memorial Lecture. Circ Res. 2012;110:755–763. doi: 10.1161/CIRCRESAHA.111.256768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaves M, Sontag ED, Dinerstein RJ. Steady-states of receptor-ligand dynamics: A theoretical framework. J Theor Biol. 2004;227:413–428. doi: 10.1016/j.jtbi.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Hu Y, Matkovich SJ, Hecker PA, Zhang Y, Edwards JR, Dorn GW., 2nd Epitranscriptional orchestration of genetic reprogramming is an emergent property of stress-regulated cardiac microRNAs. Proc Natl Acad Sci U S A. 2012;109:19864–19869. doi: 10.1073/pnas.1214996109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matkovich SJ, Hu Y, Dorn GW., 2nd Regulation of cardiac microRNAs by cardiac microRNAs. Circ Res. 2013;113:62–71. doi: 10.1161/CIRCRESAHA.113.300975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matkovich SJ, Hu Y, Eschenbacher WH, Dorn LE, Dorn GW., 2nd Direct and indirect involvement of microRNA-499 in clinical and experimental cardiomyopathy. Circ Res. 2012;111:521–531. doi: 10.1161/CIRCRESAHA.112.265736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matkovich SJ, Van Booven DJ, Eschenbacher WH, Dorn GW., 2nd RISC RNA sequencing for context-specific identification of in vivo microRNA targets. Circ Res. 2011;108:18–26. doi: 10.1161/CIRCRESAHA.110.233528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: Practical considerations. Nat Rev Genet. 2002;3:391–397. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]