FIG 7 .
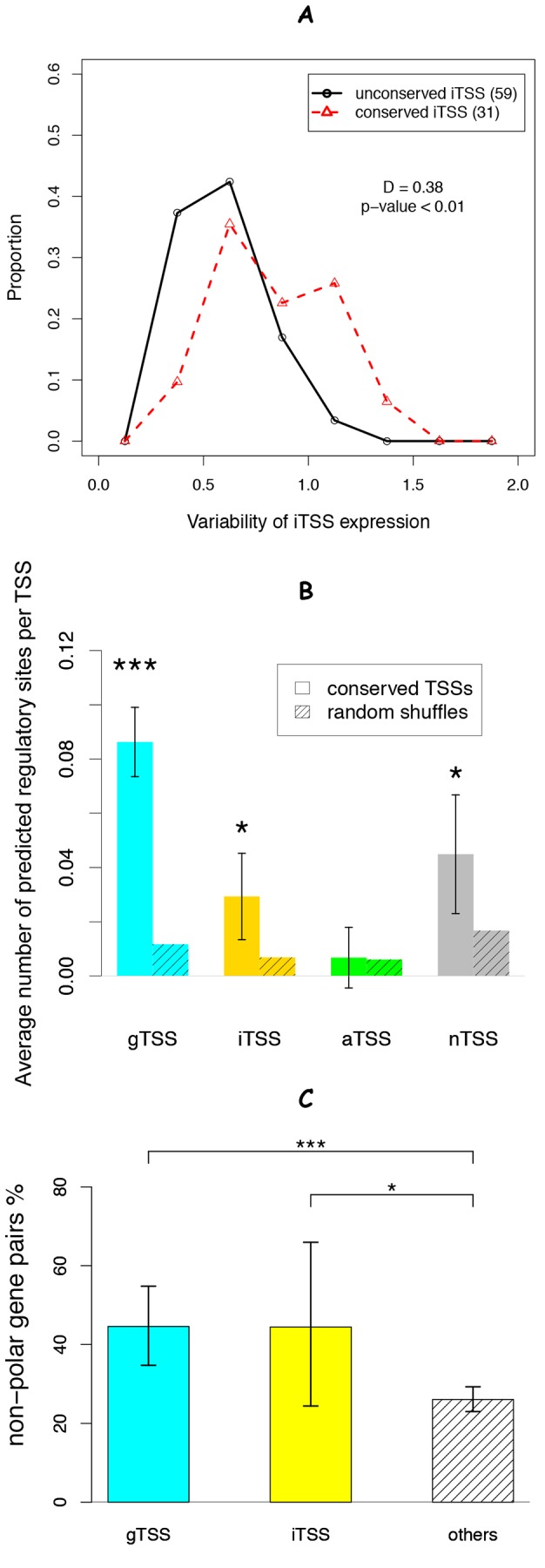
Evidence that conserved sense internal TSSs (iTSSs) are functional. (A) The distribution of the standard deviations of expression due to iTSSs across 74 conditions, as estimated from the difference between gene expression downstream and upstream of a particular iTSS. (B) The average number of predicted regulatory sites within the −50/+10 region of each group. We extracted the position weight matrixes of nine regulatory factors (ArgR, Crp, Fnr, FUR, HexR, LexA, NarP, PsrA, and TyrR) from RegPrecise (35) and scanned for hits with eight or more bits. The significance of the difference from randomly shuffled sequences was estimated by using a paired Student’s t test. (C) The percentage of operon gene pairs that lacked polar effects (upstream gene fitness greater than −0.5 and downstream gene fitness less than −1.5) in at least one experiment for mutant fitness (34). The difference between pairs with internal gTSSs, pairs with iTSSs, or other pairs was tested by using Fisher’s exact test. Error bars indicate 90% confidence intervals. *, P < 0.05; ***, P < 0.001.
