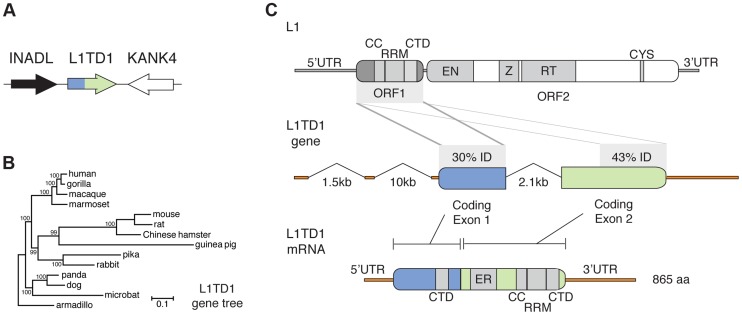
Figure 1. The mammalian L1TD1 gene was born from a tandem insertion of L1 ORF1.
A. Genomic context of L1TD1. Single-copy genes INADL and KANK4 flank L1TD1 in human, mouse and dog genomes; this shared syntenic arrangement helped us identify L1TD1 orthologs in other mammalian genomes. B. L1TD1 evolved according to the accepted species tree. We aligned L1TD1 nucleotide sequences and generated a maximum-likelihood phylogeny (see Materials and Methods). Bootstrap values show the percentage of 1000 replicates in which descendent taxa cluster together, and the scale bar shows substitutions per site according to the GTR+I+G evolutionary model. C. L1TD1 comprises two L1-ORF1p-like regions. L1 elements encode an approximately 6.5 kb transcript containing two open reading frames. L1 ORF1 encodes a protein, ORF1p, with RNA-binding and chaperone activity. ORF1p contains a coiled-coil motif (CC), a RNA-recognition motif (RRM), and a C-terminal domain (CTD). ORF2 encodes a protein with endonuclease and reverse transcription enzymatic functions. Sequence identity demonstrates that L1TD1 was formed from the domestication of two copies of ORF1 from L1. The two copies may derive from independent insertions or from duplication after a single insertion. Human coding exon 1 and coding exon 2 share 30% and 43% amino acid identity, respectively, with ORF1p of human L1.3. In coding exon 1, only the CTD is conserved, while in coding exon 2, the CC, RRM, and CTD are all conserved. Coding exon 2 also contains a variable length glutamic acid-rich region (ER). After splicing, the human L1TD1 transcript is 3849 nucleotides in length and encodes a single 865 amino acid protein product.