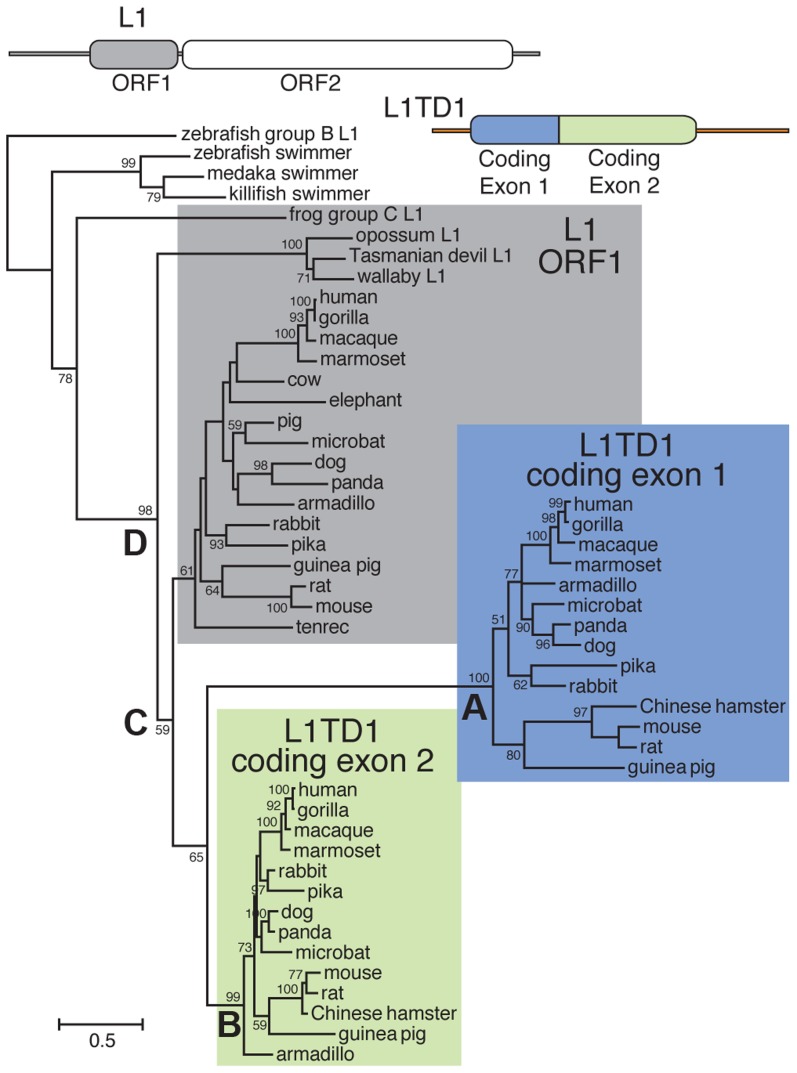
Figure 2. Phylogenetic tree of representative L1-ORF1p sequences and mammalian orthologs of the two L1TD1 ORF1p-like regions.
Predicted protein sequences of L1 ORF1p and the ORF1p-like regions encoded by each L1TD1 exon were aligned and this alignment was used to generate a maximum-likelihood tree (see Materials and Methods). Bootstrap values show percentage of 1000 replicate trees in which the descendent taxa clustered together (only values >50% shown). The scale shows the number of substitutions per site. The tree was rooted using fish swimmer elements as outgroups. The N-terminal L1TD1 ORF1p-like sequences cluster together with a high bootstrap value (node A), as do the C-terminal L1TD1 ORF1p-like sequences (node B), confirming that we have identified true L1TD1 orthologs and that the double ORF1p structure arose just once since the divergence of placental mammals and has not been subject to gene conversion between the two exons since. The L1TD1 N-terminal and C-terminal clades branch off from placental mammal L1-ORF1p sequences (node C) after marsupial and placental mammal L1-ORF1p sequences diverged (node D). The tree does not help to distinguish whether L1 ORF1 was independently domesticated twice, or just once with a subsequent genomic tandem duplication. Extensive sequence divergence between paralogous ORF1p-like sequence means that deep nodes of the tree are poorly resolved. Nonetheless, the tree supports our model in which both L1TD1 exons were born after marsupials and placental mammals diverged.