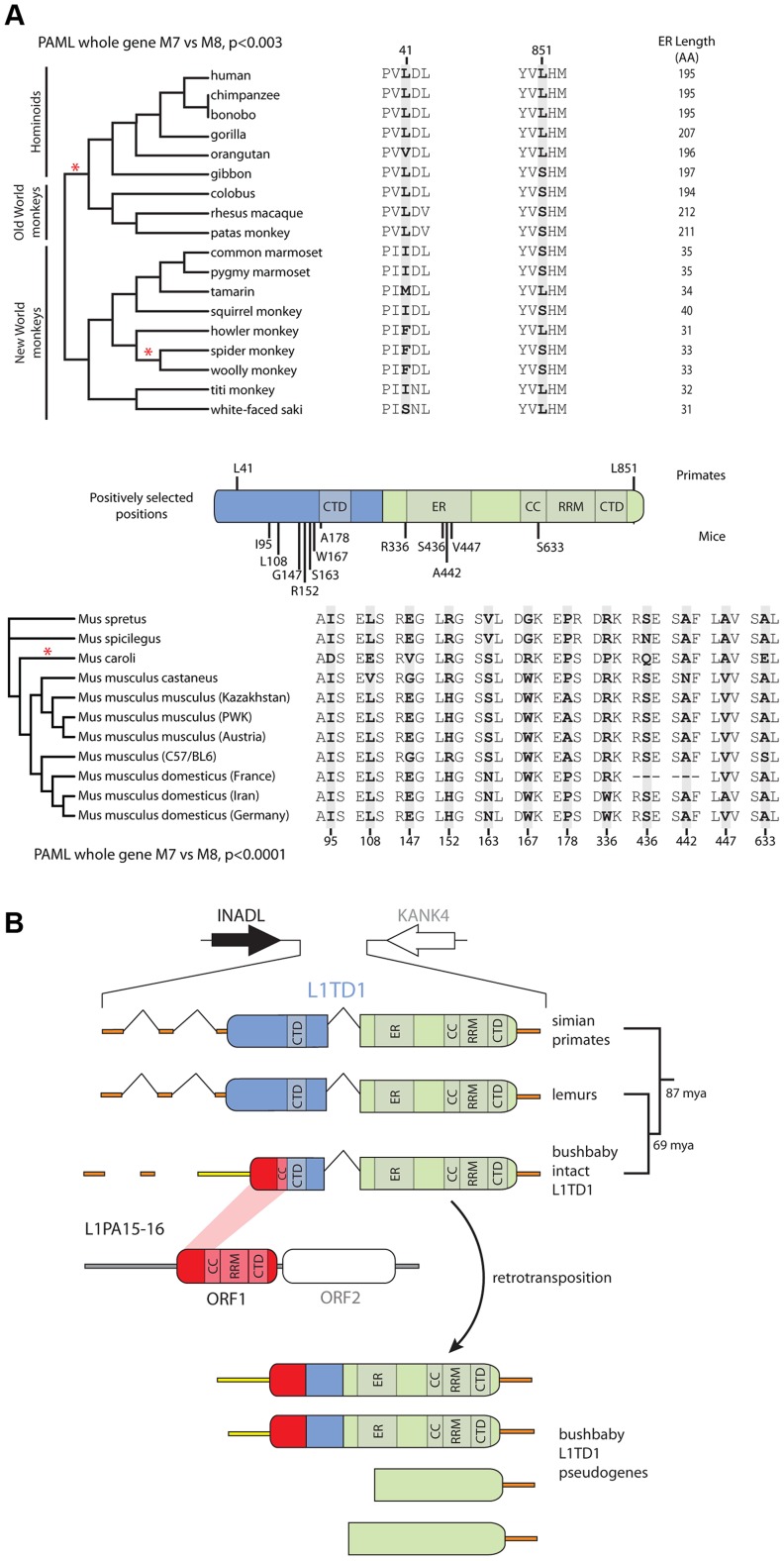
Figure 4. Novelty in L1TD1 of primates and mice.
A. Site-specific PAML analyses reveal a signature of positive selection in L1TD1. The labels on the annotated schematic indicate positions that are highly likely to be evolving under positive selection (P>90%) according to PAML NSsites (Table 1) in primates and mice (above and below the gene diagram, respectively). A species tree of primates or an L1TD1 gene tree of mice, shows branches with statistically significant episodic diversifying selection (p<0.05) according to HyPhy's Branch-site REL (marked with a red asterisk). To the right of each tree, the amino acids found at each positively selected position are shown, along with the length of the glutamic acid-rich region in each primate. Position numberings are based upon the human and M. musculus (C57/BL6) sequences. B. The L1TD1 gene of bushbaby has acquired a novel 5′ end of coding exon 1 through the insertion of a portion of a L1 element from the L1PA15-16 class (shown in red). The gene retains high sequence conservation with L1TD1 of lemurs and simian primates across the latter half of its first coding exon and all of its second coding exon (shown in blue and green). This insertion is unique amongst all the species we have examined, and is not evident in lemur genomes (mouse lemur or aye-aye). Elsewhere in the genome, bushbaby contains at least two complete and two partial processed L1TD1 pseudogenes that allowed us to infer the structure of the active L1TD1 gene.