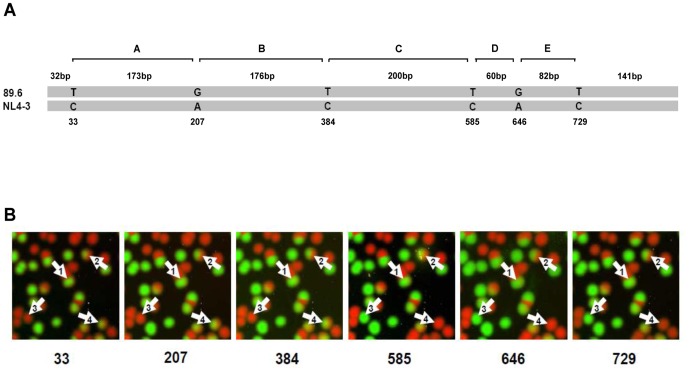
Figure 1. Detection of PCR-mediated recombinants by PASS.
(A) Nucleotides used for linkage analysis to identify recombinants are indicated. A partial pol gene (870 bp) was amplified. Nucleotides that are distinct at six positions between 89.6 and NL4-3 are shown. The regions between two neighbor nucleotides are named as A through E and the genetic distances between them are shown. (B) Linkage analysis of nucleotides at six positions by PASS. The polonies in the same PASS gel were probed by six sequential SBEs to identify recombinants. Each image represents the results from one SBE. The sequencing primers were named according to the base positions and are indicated at the bottom of the image. Each spot represents an amplicon from a single DNA molecule. The bases in 89.6 were detected by SBE with Cy5-labeled nucleotides (red) and the bases in NL4-3 were detected by SBE with Cy3-labeled nucleotides (green). The numbered arrows indicate linkage analysis results from different double-stranded DNA molecules: (1), homoduplex without recombination; (2), heteroduplex without recombination; (3), homoduplex with a recombination breakpoint between nt384 and nt585; (4), heteroduplex with a recombination breakpoint between nt384 and nt585.