Figure 1.
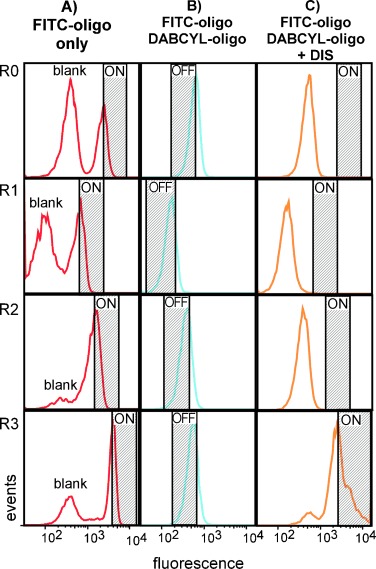
Progress of the LN8 DIS selection by flow cytometry. A) Bead-bound LN8 library annealed to the FITC oligonucleotide to determine the positive gates (“ON′′) and to confirm the presence of unlabeled beads indicating that the emulsion remained intact during PCR (”Blank“). B) Following hybridization of the FITC- and DABCYL-oligonucleotides, beads were quenched and sorted by using the hashed gate (”OFF“) to remove false-positive events. C) Positive selection was initiated by the addition of 200 μm DIS, and beads were sorted for positive events falling in the hashed gate (”ON′′), which was set based on the data collected in A). R0 data is derived from the initial LN8 library, R1 from the first enriched library, etc. Variations in the position of both the “ON” and “OFF” gates were due to day-to-day differences encountered on the flow cytometer and highlight the need for running portions of each individual round of selection prior to each sort in order to accurately define both of these states on any given day.
