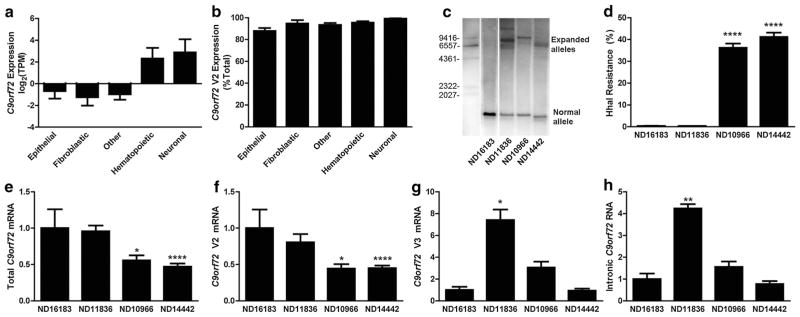
Fig. 3.
C9orf72 methylation inhibits expression of mutant RNA. a ENCODE CAGE-seq quantification of C9orf72 mRNA. The total number of C9orf72 sequence tags was normalized for number of total sequence reads, shown as mean log2 transformed tags per million (TPM) ± SE. Cell lines were divided into different cell lineages as labeled, with other representing various mesenchymal and embryonic stem cell lineages. b ENCODE CAGE-seq quantification of C9orf72 V2 mRNA relative to total C9orf72 mRNA expression. The number of V2 tag sequences was normalized to total C9orf72 tag sequences, shown as mean % of total ± SE. c Southern blot of LCL DNA from non-expanded (ND16183) and expanded (ND11836, ND10966 and ND14442) cultures using a probe specific for C9orf72 that recognizes the normal allele (bottom) and expanded alleles (top). Molecular weight markers are shown as indicated. d HhaI resistance from non-expanded (ND16183) and expanded (ND11836, ND10966 and ND14442) LCLs, shown as mean + SE. One-way ANOVA: p < 0.0001. ****p < 0.0001 relative to ND16183. Each cell line was measured in triplicate. e–h RT-qPCR quantification shown as mean + SE (n = 3–5) for total mRNA (e), V2 mRNA (f), V3 mRNA (g) and intronic RNA (h) from control (left) or expanded (right) lymphoblast cells. *p < 0.05, **p < 0.01, ****p < 0.0001