Figure 4.

Predicted binding energy matrices for Egr1 and Hnf4a. A and B are predicted PBEMs for Egr1 and Hnf4a, respectively, by BayesPI2 energy-independent model. C and D are predicted PBMEs for Egr1 and Hnf4a, respectively, by BayesPI2 energy-dependent model including dinucleotide interdependence. Figure 4E and 4F are predicted energy-dependent matrices of dinucleotide interactions (i.e.  where
where 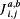 is the dinucleotide-dependent energy correction) for Egr1 and Hnf4a, respectively, by BayesPI2 energy-dependent model. In E and F, the dark and the light gray color represent the high and the low binding energy-level, respectively.
is the dinucleotide-dependent energy correction) for Egr1 and Hnf4a, respectively, by BayesPI2 energy-dependent model. In E and F, the dark and the light gray color represent the high and the low binding energy-level, respectively.
