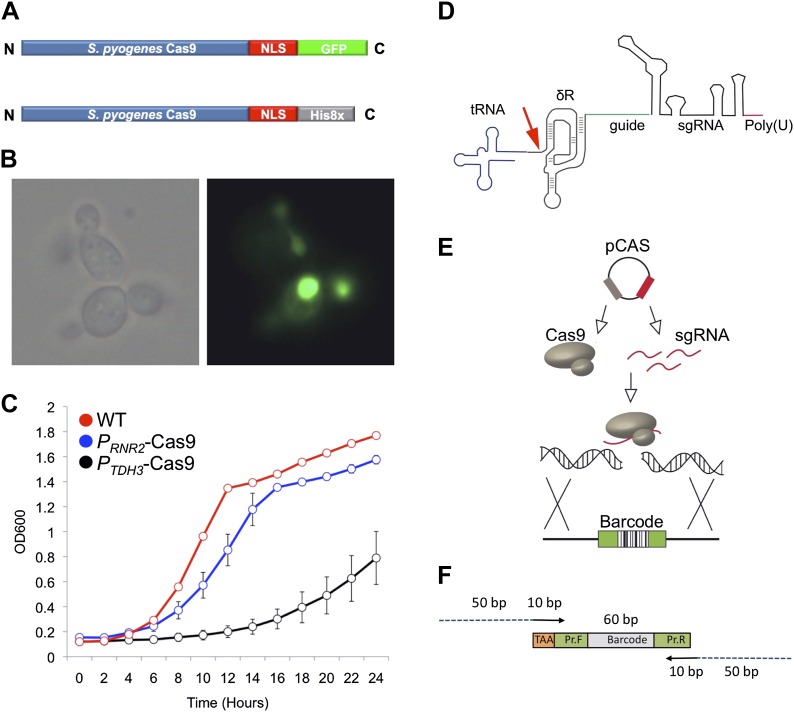
Figure 2. The engineered CRISPRm platform.
(A) Cas9 construct used for nuclear localization experiments. The S. pyogenes Cas9 protein was tagged with a C-terminal nuclear localization motif and a green fluorescent protein (GFP) (green). For genome editing experiments, the S. pyogenes Cas9 protein was tagged with a C-terminal nuclear localization motif and a Histidine affinity tag (gray). (B) Cellular localization of Cas9-GFP in exponentially growing S. cerevisiae cells. The Cas9-GFP protein was expressed from the TDH3 promoter in this experiment. Left, bright field image; right fluorescence microscopy. (C) Growth profiles of yeast expressing Cas9 from a strong promoter PTDH3 (black) or a weaker promoter PRNR2 (blue) relative to wild type (red). (D) The mature sgRNA contains a 5′ hepatitis delta virus (HDV) ribozyme (δR, brown), 20mer target sequence (green), sgRNA (black) and RNA polymerase III terminator (red). The RNA polymerase III promoter tRNA (blue) is catalytically removed by the HDV ribozyme (red arrow). (E) Schematic of yeast Cas9 targeting. Cas9 and sgRNA are expressed from a single plasmid, form a complex, and cleave targeted genomic DNA, which is repaired using a barcoded oligonucleotide. (F) The linear barcoded repair DNA molecule. Each repair DNA contains a 5′ TAA stop codon (gold), a forward primer sequence (green), a unique molecular barcode (gray), and a 3′ reverse primer (green) (Giaever et al., 2002). Barcodes are amplified using a forward primer that contains 50 bp of homology (blue) to the genome targeting site and a reverse primer that contains 50 bp of homology to the genome targeting site. The 50 bp of genomic targeting sequence are each 10 bp proximal to the PAM motif, resulting in a 20 nt deletion and barcode oligonucleotide integration.