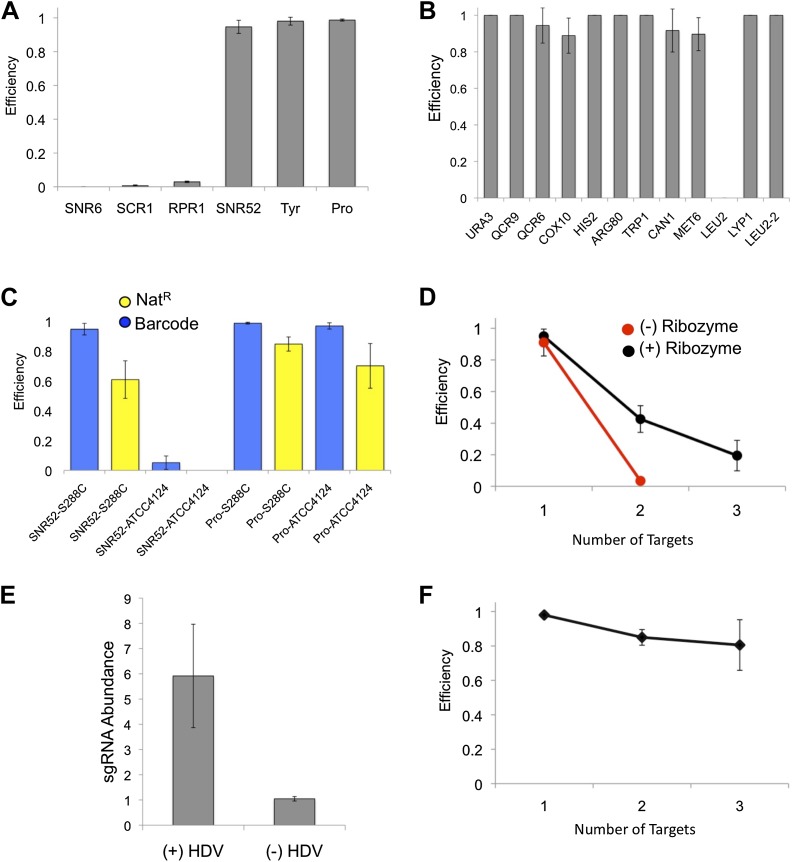
Figure 3. CRISPRm barcode insertion in yeast cells.
(A) Targeting efficiency at the URA3 locus in diploid S288C yeast using RNA polymerase III promoters (x axis) to drive the expression of the sgRNA. (B) Targeting efficiency measured across 11 loci in 10 genes in diploid cells of yeast strain S288C, using the tRNATyr promoter. (C) Single fragment barcode integration (blue) and three-fragment NatR cassette integration (yellow) efficiency in S288C diploid and ATCC4124 polyploid strains. For each experiment the promoter and strain are indicated as promoter-strain (e.g., SNR52-S288C). (D) Efficiency of multiplex insertion of barcoded DNA in diploid yeast cells with the 5′ HDV ribozyme (black) and without the 5′ HDV ribozyme (red). Triplex targeting without the 5′ HDV ribozyme was not tested. The tRNATyr promoter was used in these experiments. (E) The addition of a 5′ HDV ribozyme increases the intracellular levels of sgRNA by sixfold. The RT-qPCR experiments were carried out in biological triplicate, with the mean and standard deviation shown. (F) Efficiency of targeting one (URA3), two (URA3, LYP1) and three (URA3, LYP1 and COX10) in haploid S288C yeast cells. The tRNATyr promoter was used for sgRNA expression.