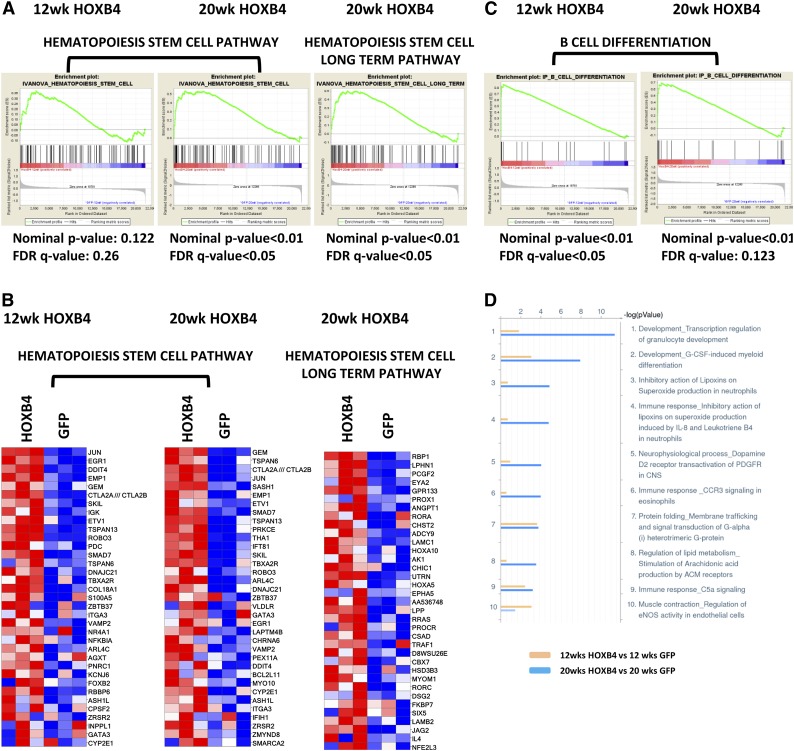
Figure 4.
GSEA and MetaCore pathway analysis of HOXB4 responsive genes. (A) GSEA analyses are shown for HOXB4 regulated genes identified for 2 different HSC pathways. LSK cell samples were obtained from transplanted mice at 12 and 20 weeks after transplant. For each GSEA, the nominal P value and FDR q-value are shown below each pathway graph. Statistical significance was validated for both pathways in 20-week samples. (B) Heatmap gene expression changes in individual gene probe sets within the Hematopoiesis Stem Cell pathway and Hematopoiesis Stem Cell Long Term pathway in LSK cells analyzed at 12 and 20 weeks after transplant. Each column represents an independent experiment within the indicated group. Red boxes represent upregulated genes and blue boxes represent downregulated genes. (C) GSEA analysis B-cell differentiation pathway genes altered by HOXB4 expression in 12- and 20-week LSK samples. Statistical parameters are listed below each analysis. (D) Pathway analysis of differentially expressed probe sets using MetaCore program analysis showing the top 10 pathways regulated by HOXB4 at 12 (yellow) and 20 weeks (blue) posttransplantation. The x-axis scale shows the −10 log (P value) of each pathway. The Development_Transcription regulation of granulocyte development pathway and Development_G-CSF-induced myeloid differentiation pathway are the top 2 pathways activated by HOXB4 at 20 weeks posttransplantation.