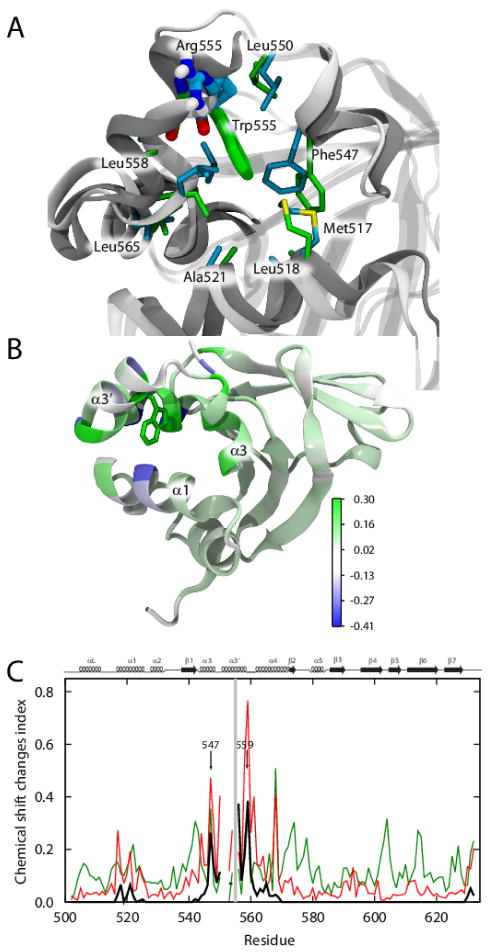
FIGURE 4. Local structural changes in the Arg555Trp mutant FAS1-4 domain caused by insertion of the tryptophan side chain into the hydrophobic cavity.
(A) The differences in side chain rotation within the cavity lined by helices α1, α3, α3′, and α4. The WT FAS1-4 domain is shown in light grey, while the Arg555Trp mutant domain is shown in dark grey. The cavity consists of the hydrophobic residues: Met517, Leu518, Ala521, Phe547, Leu550, Leu558, Leu559, and Leu565, which are here shown in cyan in the WT structure and in green in the Arg555Trp mutant structure. Arg555 (cyan) in the WT FAS1-4 domain and Trp555 (green) of the Arg555Trp mutant domain are shown in fat sticks. (B) Cartoon representation of the Arg555Trp mutant FAS1-4 domain structure showing the heavy atoms of Trp555 with green sticks, the remainder of the protein structure is coloured from blue through white to green indicating observed 1HN chemical changes. Residues Ala500, Gly501, Ala521, and Ile522 and all proline residues are coloured white to indicate missing chemical shift data and the two C-terminal residues are coloured white to avoid focus on chemical shift changes due to the C-terminal Pro634Ala mutation. (C) Observed and predicted (using shAIC [44]) chemical shift change indexes for the FAS1-4 domain showing ρobs, ρW, and 0.1* ρstruct (see definitions in the Experimental Procedures section) in red, black, and green, respectively, as a function of the residue number. Due to proline residues, some values are missing. The mutation site, residue 555 is highlighted with a grey line. The secondary structures are shown at the top of the chart for reference.