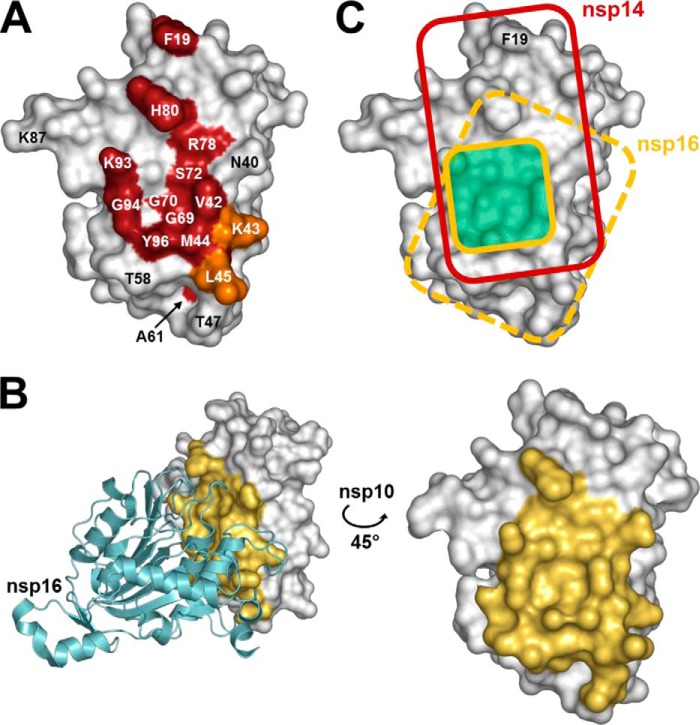
FIGURE 5.
Comparison of the nsp14 and nsp16 interaction domains on the nsp10 surface. A, schematic representation of nsp10 residues that are engaged in the nsp10-nsp14 interaction. SARS-CoV nsp10 (PDB code 2FYG (37)) is shown as a white surface representation. Residues that significantly affect the nsp10-nsp14 interaction when mutated are colored in red (>50% decrease of BRET values, binding affinity, and ExoN activity). Residues Lys-43 and Leu-45, which impair the nsp10-nsp14 interaction with a smaller effect when mutated (both binding affinity and ExoN activity ∼50% of wild type), are shown in orange. B, representation of nsp10 residues involved in the nsp10-nsp16 interaction in the structure of the complex (PDB code 2XYQ (40)). Nsp16 is shown as a schematic representation, colored in cyan. Nsp10 residues that are present within a 5 Å radius of nsp16 are depicted in yellow. C, schematic representation of nsp10 functional interacting surface with nsp14 (red rectangle) and nsp16 (dark yellow square, based on Lugari et al. (41)). The structural nsp10-nsp16 interaction surface is depicted as a dashed yellow rectangle on nsp10. The overlapping functional interaction surface is depicted in pale green. All figures were generated using PyMOL.