FIGURE 3.
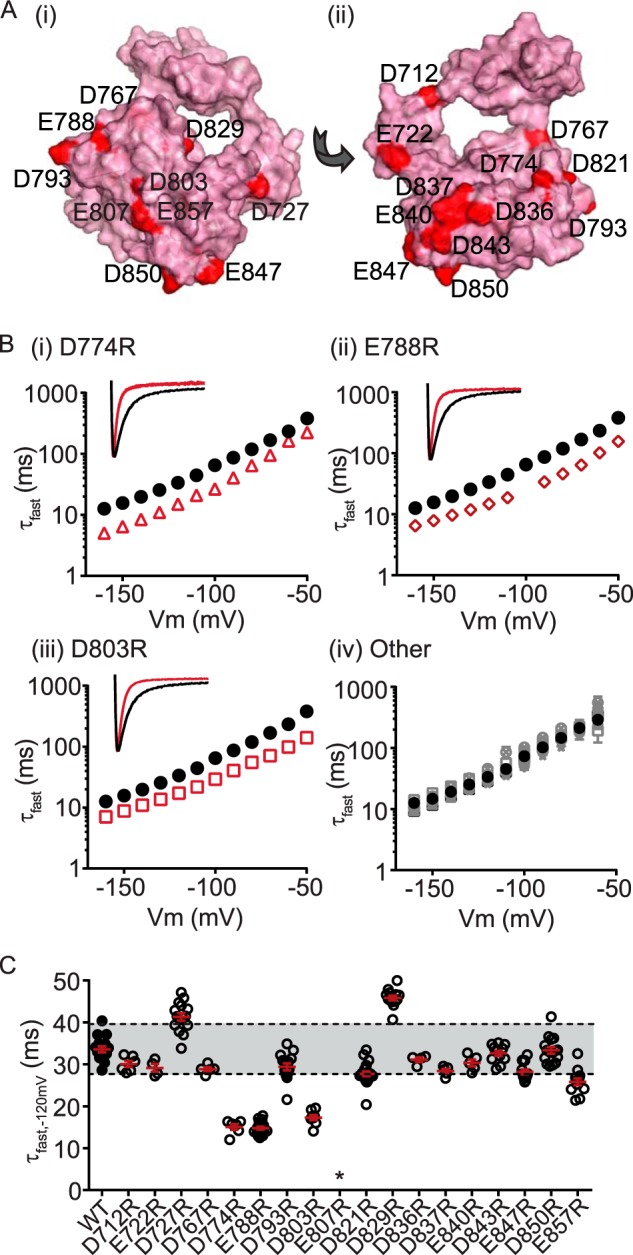
Several charged cNBH domain residues are critical for the slow deactivation kinetics of Kv11. 1 channels. A, surface representation homology model of the C-linker/cNBH domain of Kv11.1 channels showing the location of 18 negatively charged residues (red spheres). B, mean (± S.E.) rates of deactivation for the 18 negatively charged cNBH domain residues individually mutated to arginine. Three mutations exhibited substantially faster deactivation kinetics: D774R (panel (i), red triangles, n = 6), E788R (panel (ii), red diamonds, n = 21), and D803R (panel (iii), red squares, n = 8), whereas the remainder had deactivation kinetics similar to, or slower than, WT (panel (iv), gray symbols, n = 4–21). Note that error bars are included but are within the symbols. Mean ± S.E. values for τfast, τslow and the relative amplitudes of τfast to τslow for WT and all mutants are given in supplemental Table S1. Representative current traces at −120 mV for D774R, E788R, and D803R are shown in the inset. C, summary of all individual mutant effects on deactivation kinetics measured at −120 mV. Mutant perturbations were considered significant only when the mean was outside ± two standard deviations of the WT Kv11.1 (indicated by gray band). D774R, E788R, D803R, D821R, and E857R exhibited fast deactivation kinetics, whereas D727R and D829R were significantly slower than WT. * denotes that E807R did not express functional channels.
