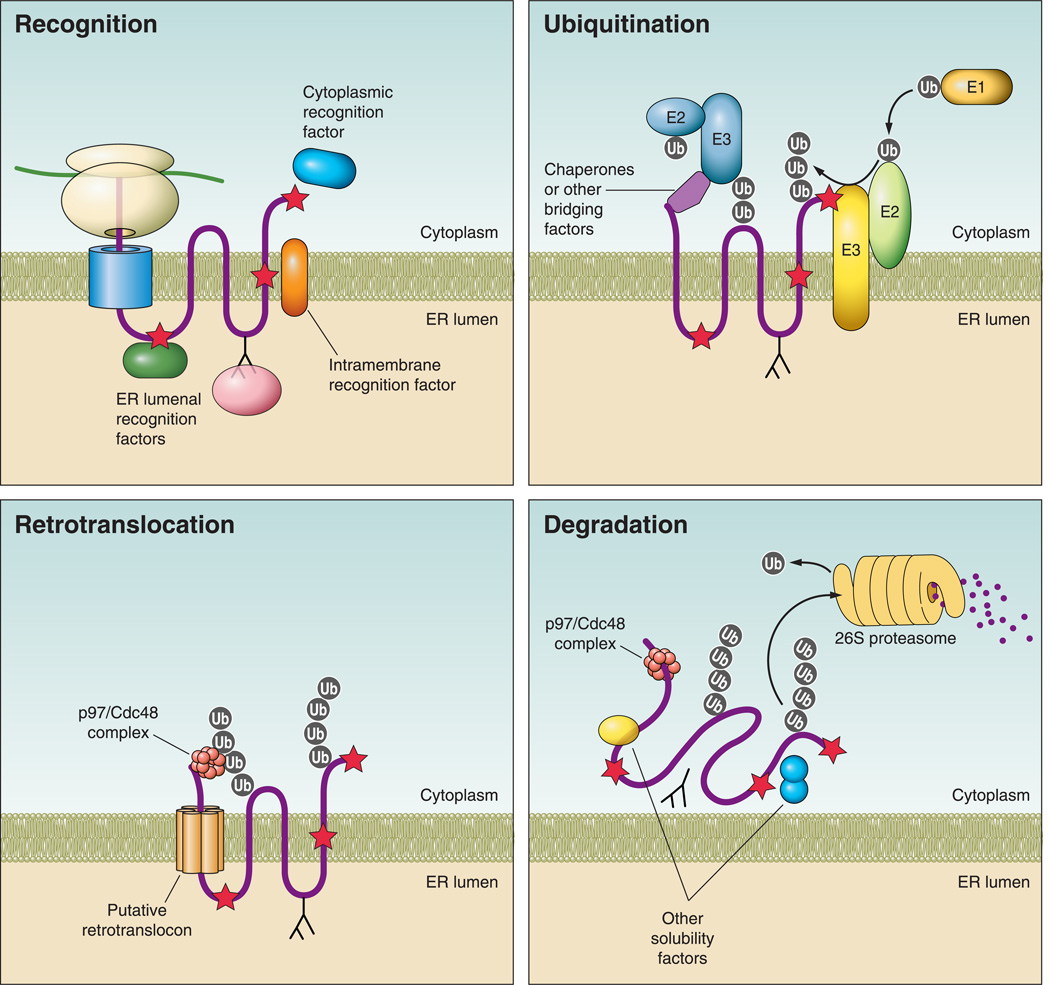
FIGURE 2. Steps in endoplasmic reticulum-associated degradation (ERAD).
Recognition: during protein synthesis and translocation, a misfolded region (red star) may reside in a protein’s cytoplasmic, ER luminal, or transmembrane domains. Recognition is mediated by ER luminal or cytoplasmic chaperones, as depicted, depending on the location of the folding lesion. For glycoproteins, lectins (pink) interact with N-glycans and in some cases they monitor the folding status of the protein. Ubiquitination: following recognition, the ubiquitination machinery is recruited to the misfolded substrate, either directly within the membrane or by interactions with cytoplasmic chaperones. A ubiquitin activating enzyme (E1) transfers ubiquitin (gray circle) to an active site cysteine in a ubiquitin conjugating enzyme (E2) in an ATP-dependent process. The ubiquitin is then transferred most commonly to a lysine residue on a client protein via a ubiquitin ligase (E3). Ubiquitination at the ER membrane can occur via cytoplasmic or ER-localized E3 ligases, both of which are shown. Retrotranslocation: for polytopic membrane proteins (pictured), retrotranslocation may occur by removal of the protein through a channel (retrotranslocon) and/or by removal of the protein and the surrounding membrane (not pictured). In either case, retrotranslocation almost always depends on the p97/Cdc48 complex, which includes Ufd1 and Npl4 and interacts with ubiquitin and misfolded regions on a substrate. p97/Cdc48 provides the mechanical force via ATP hydrolysis for substrate removal. Degradation: following retrotranslocation, misfolded proteins are ushered to the 26S proteasome and must be kept soluble to prevent aggregation. N-glycans are clipped by N-glycanse (not pictured), and ubiquitin moieties are removed by deubiquitinating enzymes either in the cytosol or in the proteasome cap. The proteasome contains three peptidase activities, trypsin-like, chymotrypsin-like, and caspase-like, which cleave proteins into short peptide fragments.