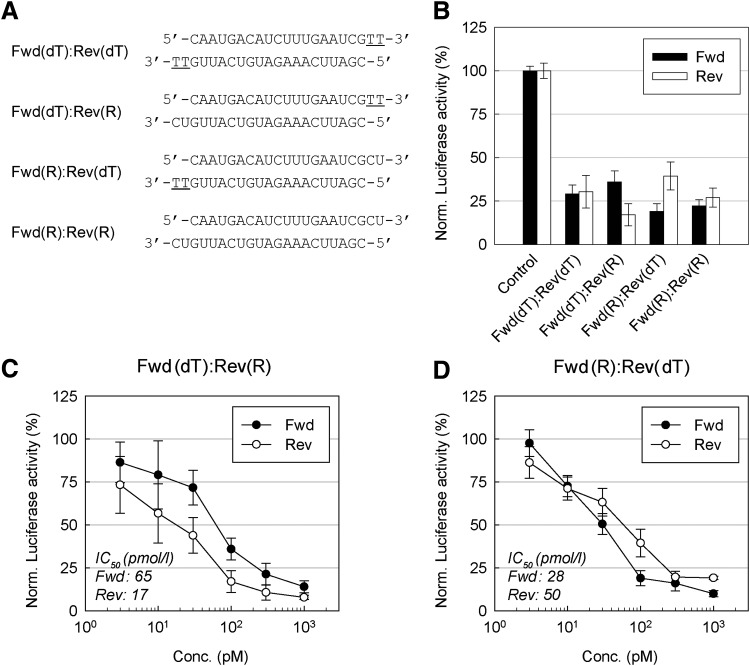
FIG. 1.
Effects of the small interfering RNA (siRNA) 3′-overhangs on the RNA interference (RNAi) activity of each strand. (A) The structures of the siRNA duplexes with symmetric or asymmetric overhangs. Underlined letters indicate DNA. (B) Reporter gene knockdown activities of the siRNAs. A549 cells were transfected with each firefly luciferase reporter construct, either with the forward (Fwd) or reverse (Rev) strand target shown in (A); Renilla luciferase expressing pRL-SV40 vector and 0.1 nM of siRNAs were used. Luciferase activity was measured 24 hours after transfection. Luciferase activities relative to mock transfection condition (reagent only) are presented as bar graphs. (C–D) Target gene knockdown activities of Fwd(dT)–Rev(R) and Fwd(R)–Rev(dT) duplexes. Diverse concentration of duplexes, reporter construct (Fwd or Rev) and pRL-SV40 vector were cotransfected into A549 cells for 24 hours and target gene knockdown activity was analyzed as described in (B). Half maximal inhibitory concentration (IC50) values for each strand are shown in the insets. All data in the graphs represent mean±standard deviation (SD) values of three independent experiments. dT, deoxythymidine; R, RNA.