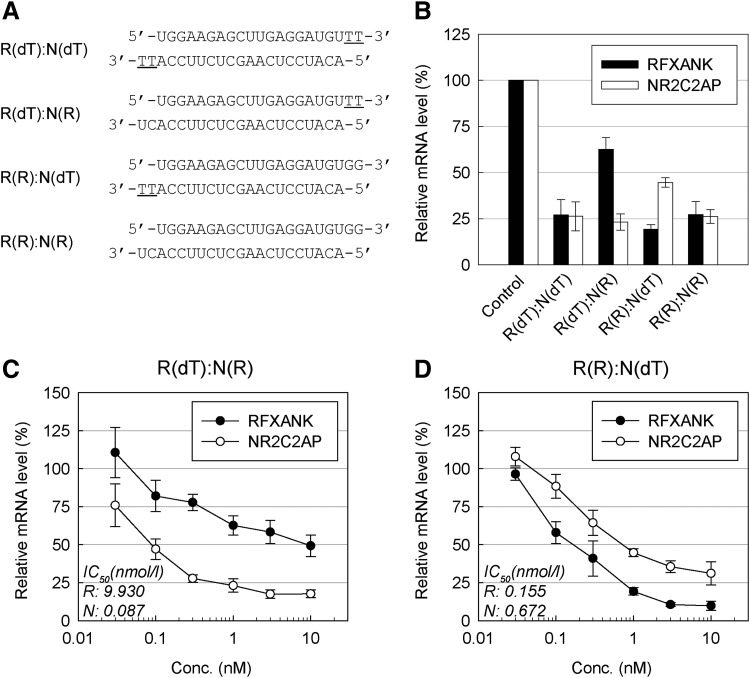
FIG. 3.
Effects of the siRNA 3′-overhangs on the RNAi activity for the endogenous sense and anti-sense transcripts. (A) The structures of the siRNA duplexes with symmetric or asymmetric overhangs. R and N represent RFXANK and NR2C2AP transcript targeting strands, respectively. Underlined letters indicate DNA. (B) Endogenous gene knockdown activities of the siRNAs. Each siRNA in (A) was transfected into A549 cells (at 1 nM of final concentration) for 24 hours, and target mRNA levels were analyzed by quantitative real-time reverse transcription polymerase chain reaction. The values plotted as “relative target mRNA level” on the y-axis in the figure were calculated as the target mRNA level divided by levels of TUBA1A (control) mRNA, and the mRNA levels relative to the mock transfection control (reagent only) are presented. (C–D) Target gene knockdown activities of R(dT)–N(R) and R(R)–N(dT) duplexes. Diverse concentrations of duplexes were transfected into A549 cells for 24 hours and target gene knockdown activity was analyzed as described in (B). IC50 values for each strand are shown in the insets. All data in the graphs represent mean±SD values of three independent experiments.