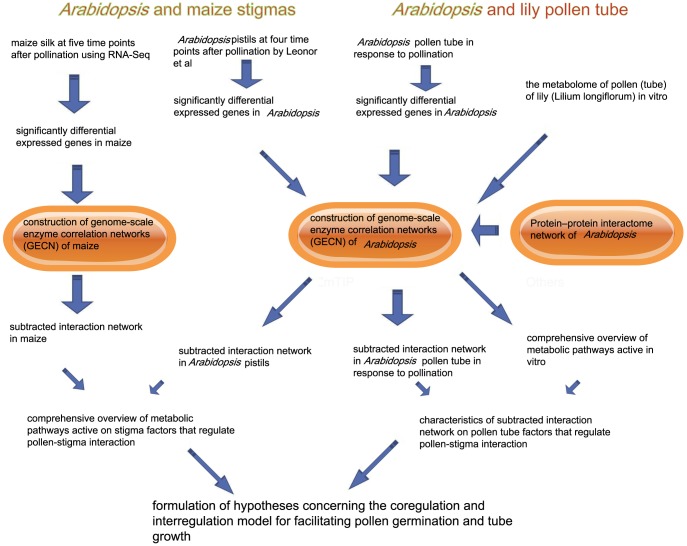
Figure 2. Schematic of “guilt by association” computational pipeline.
Metabolome of the pollen (tube) of lily (Lilium longiflorum), the significant DEGs of the Arabidopsis pollen tube and the DEGs of Arabidopsis and of maize stigmas were identified in the platform of Oracle Database and SQL (Structured Query Language). Then, metabolome of the pollen (tube) of lily, the significant DEGs that encoded enzymes of Arabidopsis and of maize were mapped to the GECN model and the protein–protein interactome network of Arabidopsis, and a sub-interaction network was constructed by the Cytoscape software. Furthermore, community analyses (NetworkAnalyzer) were used to investigate the characteristics of the systemic structure of the sub-interaction network, and to analyze transcriptional levels of genes encoding co-expressed enzymes in the consecutive steps for metabolic routes in the biological process.