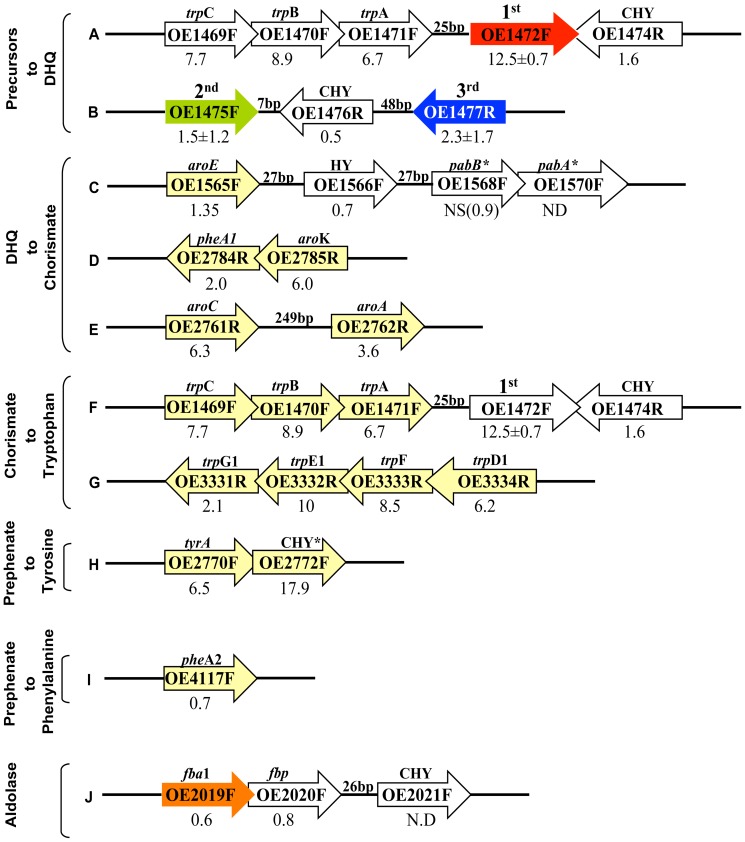
Figure 10. Genomic contexts of ORFs involved AroAA biosynthesis in H. salinarum.
Arrows show the relative positions and orientations of ORFs (but are not drawn to scale); those colored in red, green, blue or yellow represent ORFs assigned to the AroAA biosynthesis pathway while other, nearby ORFs, are uncoloured. A and B, ORFs assigned to convert precursors to DHQ. The first and second ORFs (OE1472F in red and OE1475F in green) are homologs of M. jannaschii ORFs [6]. These form part of the non-canonical part of the pathway. ORF OE1477R (in blue) is the 3rd ORF discussed in this paper. C and D, ORFs needed to convert DHQ to chorismate. F and G, the tryptophan branch. H, the tyrosine branch, I-the phenylalanine branch. J- the genomic context of ORF OE2019F, homolog to OE1472F. The numbers below the arrows represent the induction of AroAA-related genes in H. salinarum R1 cells grown in synthetic medium without AroAA relative to synthetic medium with AroAA. Induction values ≥2.0 are marked in red, and have p values of ≤10−3 unless indicated. * The two ORFs are involved in the conversion of chorismate to para-aminobenzoate, an intermediate of folate biosynthesis, NS, not significant; ND, not detected; CHY, conserved hypothetical protein; HY, hypothetical protein. ORF names and predicted functions are derived from the genome annotation at www.halolex.mpg.de.