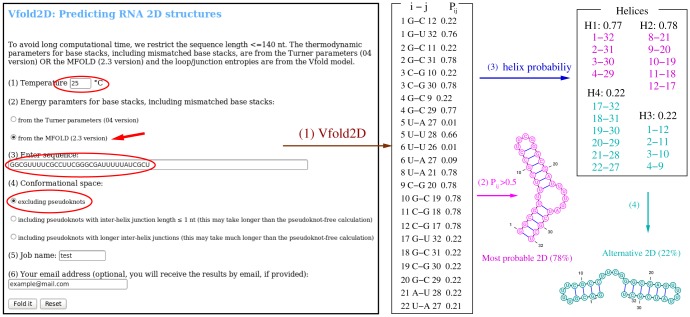
Figure 2. An example of Vfold2D prediction: the input information highlighted in the snapshot of the Vfold2D web server are the sequence (32 nts in this example), the temperature (25°C), the energy parameters used for base stacks (from MFOLD in this example) the structural type (non-pseudoknotted in this example).
(1) Vfold2D gives a list of base pair probabilities Pij (in txt format) between nucleotides i and j. For example, the probability of forming G1-C12 base pair is 0.22. (2) The most probable 2D structure is derived from the base pairs with Pij>0.5. In this example, the predicted most probable 2D structure (plotted by VARNA in the figure) has the probability of 0.78. (3) Vfold2D also predicts all the possible helices from the predicted base pair probabilities. (4) Possible alternative structures can be found from the helix and base pair probabilities (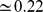 in this example).
in this example).