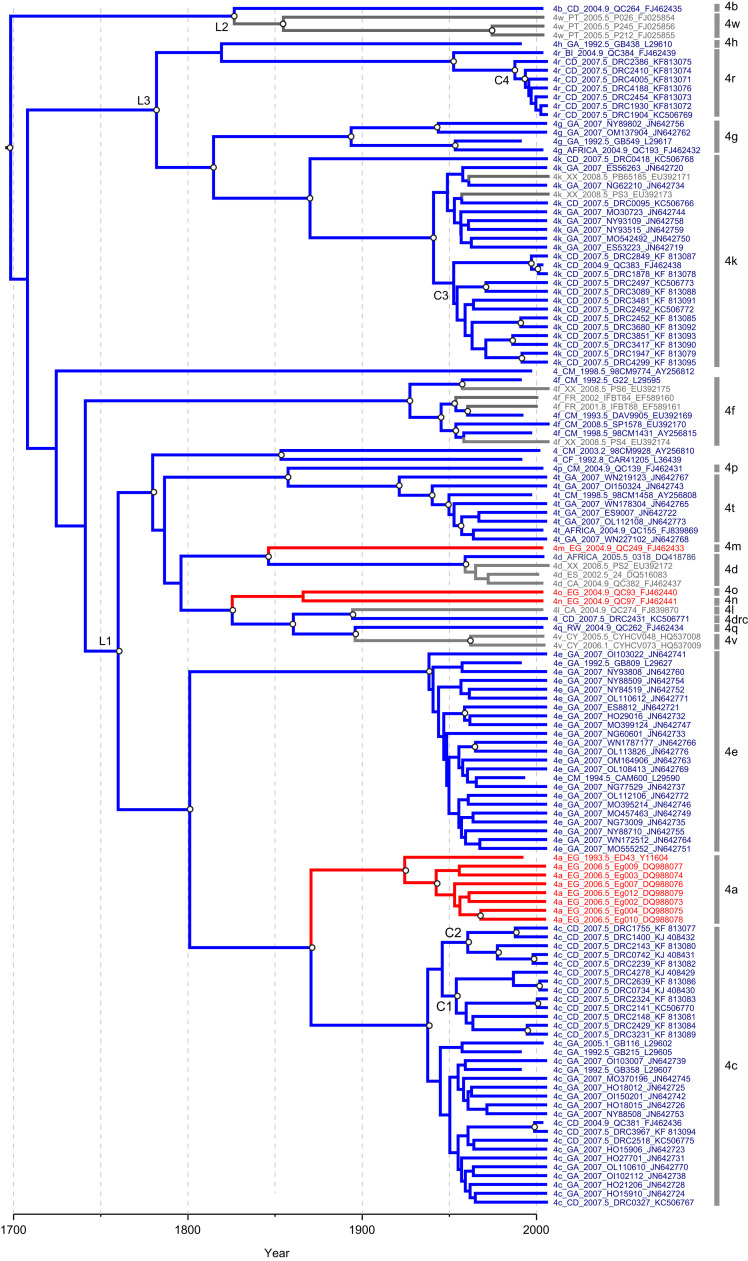
Fig. 4.
Maximum clade credibility molecular clock phylogeny, estimated from the concatenated alignment. Taxa are coloured according to location of sequence origin (blue=sub-Saharan Africa; red=Middle East and North Africa; grey=rest of the world). The locations of internal branches were inferred using parsimony and are coloured similarly. Branch lengths represent time (see scale bar at the bottom of the figure). Nodes with a posterior probability >0.9 are labelled with a white circle. Sequences are labelled as follows: subtype, sampling location using two-letter country codes (ISO 3166; see Table 4), sampling date, isolate name, accession number. XX represents an unknown location. Subtypes are indicated on the right side of the diagram. The three intra-genotypic lineages discussed in the main text are labelled L1, L2, and L3. The four clusters of samples obtained in this study discussed in the main text are labelled C1, C2, C3, and C4.