Abstract
• Premise of the study: Hyb-Seq, the combination of target enrichment and genome skimming, allows simultaneous data collection for low-copy nuclear genes and high-copy genomic targets for plant systematics and evolution studies.
• Methods and Results: Genome and transcriptome assemblies for milkweed (Asclepias syriaca) were used to design enrichment probes for 3385 exons from 768 genes (>1.6 Mbp) followed by Illumina sequencing of enriched libraries. Hyb-Seq of 12 individuals (10 Asclepias species and two related genera) resulted in at least partial assembly of 92.6% of exons and 99.7% of genes and an average assembly length >2 Mbp. Importantly, complete plastomes and nuclear ribosomal DNA cistrons were assembled using off-target reads. Phylogenomic analyses demonstrated signal conflict between genomes.
• Conclusions: The Hyb-Seq approach enables targeted sequencing of thousands of low-copy nuclear exons and flanking regions, as well as genome skimming of high-copy repeats and organellar genomes, to efficiently produce genome-scale data sets for phylogenomics.
Keywords: genome skimming, Hyb-Seq, nuclear loci, phylogenomics, species tree, target enrichment
The importance of incorporating low-copy nuclear genes in phylogenetic reconstruction is well-recognized, but has largely been constrained by technical limitations (Zimmer and Wen, 2013). These data are essential for reconstructing the evolutionary history of plants, including understanding the causes of observed incongruities among gene trees that arise from incomplete lineage sorting and introgressive hybridization. The combination of solution hybridization for target enrichment of specific genomic regions and the high sequencing throughput of current platforms (e.g., Illumina) provides the opportunity to sequence hundreds or thousands of low-copy nuclear loci appropriate for phylogenetic analyses in an efficient and cost-effective manner (Cronn et al., 2012; Lemmon and Lemmon, 2013). Most efforts to date for targeted sequencing of plant genomes for phylogenetics have been directed at the plastome (e.g., Parks et al., 2012; Stull et al., 2013). Recently, conserved orthologous sequences in Asteraceae (Chapman et al., 2007) were obtained via target enrichment for phylogenomics (Mandel et al., 2014).
Methods have been developed to target highly or ultra-conserved elements (UCEs) in animal genomes (Faircloth et al., 2012; Lemmon and Lemmon, 2013; McCormack et al., 2013). However, UCEs in plants are nonsyntenic, and are hypothesized to have originated via horizontal transfer from organelles or de novo evolution (Reneker et al., 2012). Whatever their origin, their potential for nonorthology among species makes them unsuitable as phylogenetic markers in plants. The frequency of polyploidy throughout angiosperm evolution (Jiao et al., 2011) also impedes obtaining a large set of conserved orthologous single-copy loci transferable across plant lineages, which in combination with the lack of orthologous UCEs, means that design of targeted sequencing strategies for plant nuclear genomes will necessarily be lineage-specific.
Here we present Hyb-Seq, a protocol that combines target enrichment of low-copy nuclear genes and genome skimming (Straub et al., 2012), the use of low-coverage shotgun sequencing to assemble high-copy genomic targets. Our protocol improves upon the methods of Mandel et al. (2014) by (1) utilizing the genome and transcriptome of a single species for probe design, which makes our approach more generally applicable to any plant lineage; (2) obtaining additional data from the procedure through combination with genome skimming; and (3) developing a data analysis pipeline that maximizes the data usable for phylogenomic analyses. Furthermore, we assemble sequences from the flanking regions (the “splash zone”) of targeted exons, yielding noncoding sequence from introns or sequence 5′ or 3′ to genes, which are potentially useful for resolving relationships at low taxonomic levels. We demonstrate the feasibility and utility of Hyb-Seq in a recent, rapid evolutionary radiation: Asclepias L. (Apocynaceae). Target enrichment probes were designed using the A. syriaca L. draft genome and transcriptome sequences in concert to identify nuclear loci of sufficient length (>960 bp) for robust gene tree reconstruction with a high probability of being single copy (>10% divergence from all other loci in the target genome). We also demonstrate the utility of the Asclepias data for phylogenomic analysis and explore the utility of the probes for Hyb-Seq in another genus of the same subtribe, Calotropis R. Br. (Asclepiadineae), and a more distantly related genus, Matelea Aubl. (Gonolobineae). The Hyb-Seq approach presented here efficiently obtains genome-scale data appropriate for phylogenomic analyses in plants, and highlights the utility of extending genomic tools developed from a single individual for use at deeper phylogenetic levels.
METHODS AND RESULTS
Targeted enrichment probe design
An approach was developed for Hyb-Seq probe design (Table 1) in Asclepias utilizing a draft assembly of the A. syriaca nuclear genome (Weitemier et al., unpublished data), which was assembled using Illumina paired-end data from libraries with insert sizes of 200 and 450 bp and a k-mer size of 79 in ABySS v. 1.3.2 (Simpson et al., 2009) with reads of plastid or mitochondrial origin removed prior to assembly. A transcriptome of A. syriaca leaf and bud tissue (Straub et al., unpublished data) was assembled de novo using Trinity RNA-Seq v. r20131110 (Grabherr et al., 2011) and refined using transcripts_to_best_scoring_ORFs.pl (included with Trinity). Probe design was based on data from the draft genome, which was combined with transcriptome assembly data to target the exons of hundreds of low-copy loci. Contigs from the draft nuclear genome were matched against those sharing 99% sequence identity from the transcriptome using the program BLAT v. 32 × 1 (Kent, 2002). BLAT accommodates large gaps in matches between target and query sequences, and is suitable for matching the exon-only sequence of transcripts with the intron-containing genomic sequence, allowing the locations of potential intron/exon boundaries to be identified. Additionally, in an effort to prevent loci present in multiple copies within the genome from being targeted, only those transcripts with a single match against the genome were retained. To prevent probes from enriching multiple similar loci, any targets sharing ≥90% sequence similarity were removed using CD-HIT-EST v. 4.5.4 (Li and Godzik, 2006). The remaining transcriptome contigs were filtered to retain only those containing exons ≥120 bp totaling at least 960 bp. The lower cutoff was necessary to provide sufficiently long sequences for probe design (=120 bp), and the upper cutoff was chosen to exclude short loci less likely to include phylogenetically informative sites. Of the loci that passed filtering, all of those matching (70% sequence identity over 30% of its length) a previously characterized putative ortholog from Apocynaceae (expressed sequence tags [ESTs] from Catharanthus roseus (L.) G. Don; Murata et al., 2006), the asterids (COSII; Wu et al., 2006), or four nonasterid angiosperms (Duarte et al., 2010) were retained (1335 exons in 350 loci). Additional loci that passed filtering were added to the set of targeted loci until the total length of the target probes approached the minimum required for oligonucleotide synthesis (2050 exons in 418 loci). The final probe set also contained probes intended to generate data for other projects (157 defense-related and floral development genes and 4000 single nucleotide polymorphisms [SNPs]), which were only included here where necessary for calculations of hybridization efficiency and assembly length. Note that care should be taken during the probe design process to avoid targeting organellar sequences together with nuclear sequences, because enrichment of organellar targets will be proportional to their presence in the genomic DNA extractions used to prepare sequencing libraries and may greatly exceed nuclear targets (see Appendix S1 (44.6KB, pdf) ).
Table 1.
Hyb-Seq target enrichment probe design and bioinformatics pipeline. A script combining and detailing the steps of the probe design process, Building_exon_probes.sh, is provided in the supplementary materials (Appendix S1 (44.6KB, pdf) ).
| Steps | Description | Primary program or custom script |
| Probe design | ||
| Match | Find genome and transcriptome sequences with 99% identity. | BLATa |
| Filter | Retain single hits of substantial length. | Part of Building_exon_probes.shb |
| Cluster | Remove isoforms and loci sharing >90% identity. | CD-HIT-ESTc, grab_singleton_clusters.pyb |
| Filter | Retain loci with long exons summing to desired length. | blat_block_analyzer.pyb |
| Cluster | Remove exons sharing >90% identity. | CD-HIT-ESTc, grab_singleton_clusters.pyb |
| Short read processing and data analysis | ||
| Read processing | Adapter trimming, quality filtering | Trimmomaticd |
| Exon assembly | Reconstruct a sequence for each sample, for each exon. | YASRAe, Alignreadsf |
| Identify assembled contigs | If contig identity is unknown, identify which targeting exon(s) it corresponds to. | BLATa |
| Sequence alignment I: Collate exons | Cluster orthologous exons across samples. | assembled_exons_to_fasta.pyb |
| Sequence alignment II: Perform alignment | Align homologous bases within each exon. | MAFFTg |
| Concatenate exons | For each locus, concatenate the aligned exons. | catfasta2phyml.plh |
| Gene tree construction | For each locus, estimate the maximum likelihood gene tree. | RAxMLi |
| Species tree construction | Estimate the species tree from independent gene trees in a coalescent framework. | MP-ESTj |
New scripts written for this protocol, an example data set, and any future updates are available at https://github.com/listonlab/.
Illumina library preparation and Hyb-Seq
DNA was extracted from 10 species of Asclepias, Calotropis procera (Aiton) W. T. Aiton, and Matelea cynanchoides (Engelm. & A. Gray) Woodson (Appendix 1) using either a modified cetyltrimethylammonium bromide (CTAB) protocol (Doyle and Doyle, 1987), DNeasy (QIAGEN, Valencia, California, USA), FastDNA (MP Bio, Santa Ana, California, USA), or Wizard kits (Promega Corporation, Madison, Wisconsin, USA). Most indexed Illumina libraries were prepared as described by Straub et al. (2012). Two exceptions were A. cryptoceras S. Watson (prepared with a NEXTflex DNA barcode; Bioo Scientific, Austin, Texas, USA) and M. cynanchoides (TruSeq library preparation kit; Illumina, San Diego, California, USA). Libraries were then pooled in 11- or 12-plexes with approximately equimolar ratios (some samples included in the pools were not included in the current study). Solution hybridization with MYbaits biotinylated RNA baits (MYcroarray, Ann Arbor, Michigan, USA) and enrichment followed Tennessen et al. (2013) with approximately 350–480 ng of input DNA per pool and 12 rather than 15 cycles of PCR enrichment to decrease the production of PCR duplicates. These target-enriched libraries were then sequenced on an Illumina MiSeq at either Oregon Health Science University (version 2 chemistry) to obtain 2 × 251-bp reads or Oregon State University (version 3 chemistry) to obtain 2 × 76-bp reads. Raw Illumina data were submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRP043058).
Data analysis pipelines
Raw data were filtered for adapter sequences either by the sequencing centers, using Trimmomatic v. 0.20 or 0.30 (Bolger et al., 2014), or using custom scripts. Internal sequence barcodes were deconvoluted using bc_sort_pe.pl (Knaus, 2012). Reads were quality filtered using Trimmomatic to remove bases at read ends with qualities lower than Q20, to trim the rest of the read when average quality in a 5-bp window was <Q20, and to remove reads shorter than 36 bp following trimming. For A. cryptoceras, only read ends were trimmed to Q20. Duplicate reads were removed using the FASTX-Toolkit (Gordon, 2010). For target assembly, a reference-guided approach utilizing a pseudo-reference consisting of targeted exons separated by 200 Ns was implemented in Alignreads v. 2.25 (Straub et al., 2011). BLAT was used to identify contigs in the final assembly with sequence similarity to targeted exons. A custom script extracted the longest assembled sequence corresponding to each exon and constructed matrices for multiple sequence alignment, while adding Ns to the matrix if an exon was missing for a particular species. Exons were aligned using default settings in MAFFT v. 6.864b (Katoh and Toh, 2008). Following alignment, exons of the same gene were joined using catfasta2phyml.pl. Splash-zone sequences were not included in this analysis. The same read pools were then used for reference-guided assembly of high-copy sequences, the plastome and nuclear ribosomal DNA (nrDNA) cistrons (18S-5.8S-26S), using Alignreads. The references used for each species were generated through analysis of previously collected genome skim data of the same libraries used for this study (Straub et al., 2012; Straub et al., unpublished data). References from a different A. cryptoceras individual were used for that species and reads were retrimmed using the Trimmomatic setting described above. The M. biflora (Raf.) Woodson plastome (GenBank: KF539850.1) and the C. procera nrDNA sequence served as references for M. cynanchoides. MAFFT was used for alignment. Appendix S2 (105.3KB, pdf) provides additional details on bioinformatic analyses.
Analyses of assembled sequences
The total length of assembled sequence, numbers of targeted exons and genes assembled, amount of flanking sequence assembled from the splash zone, percentage of plastome and nrDNA cistron sequence assembled from the off-target reads based on the lengths of the reference sequences, and percent divergence from the A. syriaca exon sequences were calculated for each species. The Hyb-Seq data were also analyzed using the phyluce v. 1.4 pipeline (Faircloth et al., 2012; Faircloth, 2014) used by Mandel et al. (2014), using both the native de novo assembly option and using the contigs produced by the reference-guided assembly in Alignreads as input data (see Appendix S3 (89.6KB, pdf) for detailed methods). To demonstrate the utility of the data for phylogenomics, analyses of Asclepias and outgroup Calotropis were conducted for nuclear genes individually (excluding seven genes with terminals with all missing data), a concatenation of all nuclear genes, and whole plastomes using RAxML v. 7.3.0 (Stamatakis, 2006) with a GTR + Γ model of nucleotide substitution. One hundred and 1000 rapid bootstrap replicates were conducted for nuclear and plastid analyses, respectively. Prior to analysis, the plastome matrix was edited following Straub et al. (2012). RAxML nuclear gene trees were then used for phylogenomic analyses of all targeted loci with complete taxon sampling (n = 761) using the MP-EST species tree approach with bootstrap evaluation of clade support implemented through the STRAW webserver (Shaw et al., 2013). Targeted nuclear exon sequences, data matrices, and trees were submitted to Figshare (http://dx.doi.org/10.6084/m9.figshare.1024614). See Appendices S1 (44.6KB, pdf) and S2 (105.3KB, pdf) for detailed discussion of the protocol from probe design to data analysis.
Hyb-Seq results
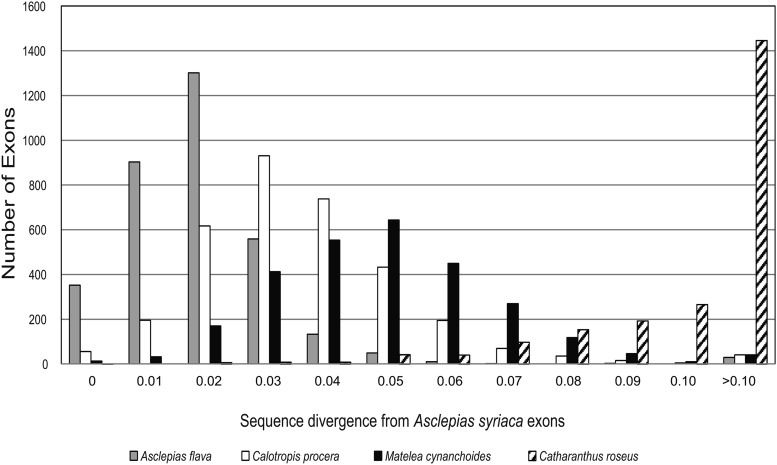
We identified 768 putatively single-copy genes (3385 exons, ca. 1.6 Mbp) meeting the criteria of sufficient length and divergence from all other genes in the genome. Of these genes, 136 genes were among asterid COSII sequences and 42 were among genes conserved across four angiosperm genomes; only 12 out of 155 possible overlapping genes were shared by both conserved sets. Enrichment, sequencing, and assembly of the targeted putatively single-copy genes was successful in Asclepias and related Apocynaceae with at least partial assembly of an average of 92.6% of exons and 99.7% of genes and a total average assembly length for all genes in the probe set of ca. 2.2 Mbp from 1.7 Mbp of targeted exons (including the defense and floral development genes; Table 2). Lower read numbers (due to unequal library pooling) resulted in reduced target capture and assembly efficiency in A. eriocarpa Benth. and A. involucrata Engelm. ex Torr. (Table 2), while a combination of lower read number and sequence divergence (average 4.5%) between Matelea and the probes is likely responsible for its somewhat lower success (Fig. 1; Table 2). In contrast, target capture in Calotropis (average 3.2% divergence from A. syriaca) was similar to Asclepias (Table 2). Given that the probes should work well up to 10% sequence divergence, this probe set is likely useful for enrichment of the targeted genes across Asclepiadoideae (Fig. 1). Extending the comparison to the more distantly related Catharanthus roseus (Gongora-Castillo et al., 2012), BLAT analysis reveals an average 12% divergence between A. syriaca exons and orthologous transcripts (Fig. 1). This result predicts that a smaller, but not insignificant, amount of sequence data could be obtained from the rest of Apocynaceae. Modification of hybridization conditions could further increase success for more divergent species (Li et al., 2013). In addition to the targeted nuclear loci, reference-guided assembly of the off-target reads yielded complete or nearly complete plastome and nrDNA cistron sequences (Table 2).
Table 2.
Success of Hyb-Seq for targeted sequencing and assembly of nuclear genes combined with genome skimming of high-copy targets in Asclepias and related species of Apocynaceae.
| Species | Readsa | Quality-filtered reads | Unique, on-target, quality-filtered reads (%)b,c | Assembly length (Mbp)b | Splash zone assembly length (Mbp)b | Single-copy gene exons assembledd | Single-copy genes assembledd | % Divergence from single-copy gene probese | % Missing data in matrix | % Completion of plastome | % Completion of nrDNA cistron |
| Asclepias cryptoceras | 1,174,294 | 1,149,278 | 746,909 (65.0) | 3.2 | 1.6 | 3349 | 768 | 0.9 | 7.4 | 99.7 | 100 |
| Asclepias engelmanniana | 1,943,370 | 1,804,956 | 523,477 (29.0) | 2.7 | 1.0 | 3359 | 767 | 0.8 | 3.6 | 97.8 | 98.3 |
| Asclepias eriocarpa | 393,048 | 384,595 | 72,200 (18.8) | 1.1 | 0.5 | 2260 | 762 | 0.9 | 69.0 | 81.9 | 94.4 |
| Asclepias flava | 1,457,860 | 1,301,608 | 397,798 (30.6) | 2.2 | 0.8 | 3313 | 768 | 1.5 | 14.7 | 98.4 | 100 |
| Asclepias humistrata | 918,608 | 843,463 | 234,502 (27.8) | 2.0 | 0.8 | 3163 | 768 | 1.0 | 27.1 | 93.1 | 97.0 |
| Asclepias involucrata | 664,820 | 645,580 | 139,407 (21.6) | 1.7 | 0.7 | 2978 | 768 | 0.9 | 41.8 | 90.5 | 99.4 |
| Asclepias masonii | 1,097,532 | 971,606 | 270,123 (27.8) | 2.1 | 0.9 | 3275 | 768 | 1.4 | 30.6 | 99.1 | 100 |
| Asclepias nyctaginifolia | 2,482,686 | 2,295,691 | 558,822 (24.3) | 2.4 | 0.8 | 3369 | 768 | 0.9 | 2.1 | 96.0 | 100 |
| Asclepias scheryi | 1,345,732 | 1,295,739 | 384,451 (29.7) | 2.4 | 0.8 | 3314 | 768 | 1.0 | 4.9 | 98.7 | 100 |
| Asclepias tomentosa | 1,248,940 | 1,111,909 | 310,020 (27.9) | 2.1 | 0.8 | 3208 | 768 | 0.9 | 26.7 | 95.2 | 99.7 |
| Calotropis procera | 1,172,456 | 1,135,014 | 380,155 (33.5) | 2.6 | 1.0 | 3287 | 768 | 3.2 | 5.0 | 96.0 | 100 |
| Matelea cynanchoides | 418,590 | 388,064 | 208,835 (53.8) | 1.7 | 0.4 | 2718 | 757 | 4.5 | n/a | 99.4 | 100 |
| Average | 1,190,419 | 1,110,625 | 352,225 (32.5) | 2.2 | 0.8 | 3133 | 767 | 1.5 | 21.2 | 95.5 | 99.1 |
Most samples were sequenced in a single MiSeq run (11-plex 2 × 251-bp version 2 chemistry) except for A. cryptoceras and M. cynanchoides, which were each sequenced in different MiSeq runs (12-plex 2 × 251-bp version 2 chemistry and 15-plex 2 × 76-bp version 3 chemistry, respectively).
These values were calculated using the entire probe set, including single-copy gene, defense and floral development genes, and SNPs.
These estimates are lower than the true overall efficiency due to quality filtering and the removal of duplicate reads. Except for A. cryptoceras and M. cynanchoides, the libraries were made with internal barcodes, which apparently contributed to suboptimal base calling and lower-quality scores, leading to apparent suboptimal target capture efficiency.
These estimates are based on a minimum 90% sequence identity to the A. syriaca probes, and are therefore conservative; especially so for C. procera and M. cynanchoides, which are expected to have higher sequence divergence.
These estimates are based on a minimum 75% sequence identity to the A. syriaca probes.
Fig. 1.
Histogram of exon sequence divergence between the species used for probe design, Asclepias syriaca, and four other species: the most divergent species of Asclepias, A. flava; another member of Asclepiadinae (Asclepiadeae: Asclepiadoideae), Calotropis procera; a member of Gonolobinae (Asclepiadeae: Asclepiadoideae), Matelea cynanchoides; and a member of a different subfamily, Catharanthus roseus (Rauvolfioideae). Note that a maximum sequence divergence of 75% was allowed for BLAT and that exons with >10% divergence were less likely to be observed in Calotropis and Matelea because they were less likely to be enriched by the probes, while the Catharanthus data were from transcriptome sequences of multiple tissues and not subject to target enrichment bias.
The data analysis pipeline presented here resulted in a data set with few missing genes for each species. In contrast, the phyluce pipeline recovered comparatively few loci for phylogenomic analysis (Table 3). Phyluce was designed for the analysis of UCE data, and its adoption for analysis of single-copy genes where multiple exons have been targeted is inappropriate because exons are often assembled on separate contigs and phyluce views multiple contigs matching a targeted locus as an indication of paralogy (see Appendix S3 (89.6KB, pdf) for further discussion). The use of reference-guided assembly in the pipeline presented here, rather than the de novo approach of phyluce, also results in a greater amount of data recovery for use in phylogenomic analyses (Table 3).
Table 3.
Number of single-copy genes recovered for phylogenomic analysis with different data analysis pipelines.
| Species | Hyb-Seq | phyluce | phyluce with Alignreads contigs |
| Asclepias cryptoceras | 768 | 16 | 145 |
| Asclepias engelmanniana | 767 | 69 | 201 |
| Asclepias eriocarpa | 762 | 10 | 23 |
| Asclepias flava | 768 | 28 | 109 |
| Asclepias humistrata | 768 | 27 | 62 |
| Asclepias involucrata | 768 | 3 | 24 |
| Asclepias masonii | 768 | 8 | 38 |
| Asclepias nyctaginifolia | 768 | 13 | 198 |
| Asclepias scheryi | 768 | 69 | 186 |
| Asclepias tomentosa | 768 | 21 | 54 |
| Calotropis procera | 768 | 84 | 203 |
| Matelea cynanchoides | 757 | 51 | 98 |
| Average | 767 | 33 | 112 |
Phylogenomics
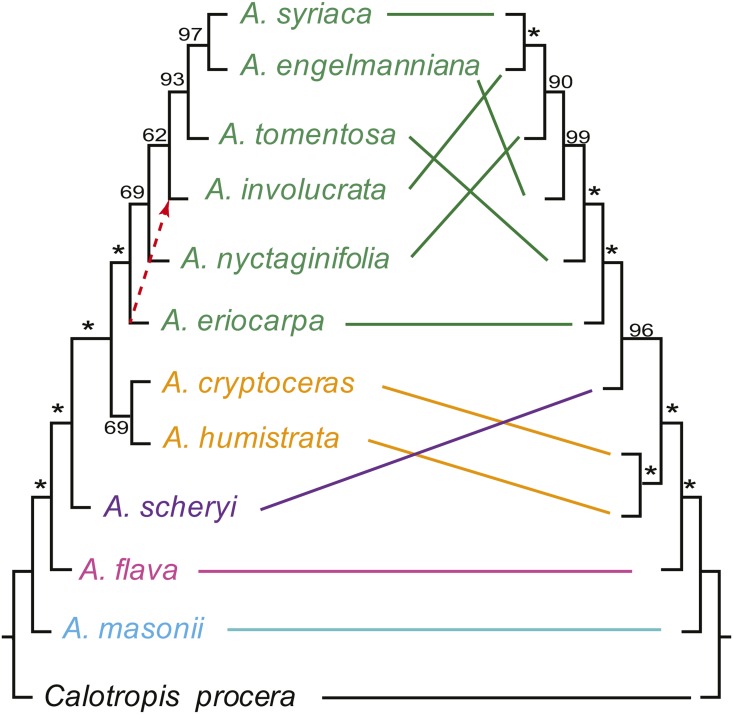
Percentage of variable sites within 768 sequence alignments ranged from 1.8% to 12.5%, with a mean of 5.9% (Appendix S4 (24.1KB, pdf) ). The concatenated data matrix was 1,604,805 bp, with 104,717 variable sites, 10,210 of which were parsimony informative. Phylogenomic analysis of the maximum likelihood gene trees for the 761 putatively single-copy genes containing information for all 12 taxa resulted in a species tree topology in which most nodes received high bootstrap support, and which differed from the concatenation species tree in the placement of A. eriocarpa (Fig. 2, left). This result highlights the importance of utilizing species tree methods and approaches for assessing clade support that take into account discordance among gene trees, because concatenation approaches can result in strongly supported, but misleading inferences of evolutionary relationships (Kubatko and Degnan, 2007; Salichos and Rokas, 2013). Maximum likelihood analysis of plastomes resulted in a resolved and well-supported phylogeny with a topology in conflict with that of the species tree, especially among temperate North American species (Fig. 2, right). Relationships in this clade estimated from noncoding plastid sequences have been shown to be at odds with expectations based on morphology (Fishbein et al., 2011).
Fig. 2.
Comparison of the species tree of Asclepias based on 761 putatively single-copy loci and the whole plastome phylogeny. The MP-EST tree is shown at left, and the difference between this topology and that recovered through an analysis of the concatenated nuclear gene data set is indicated by the red arrow. Solid lines connect each species to its placement in the plastome phylogeny (right). Values near the branches are bootstrap support values (* = 100%). Colors reflect the plastid clades of Fishbein et al. (2011): temperate North America (green), unplaced (orange), highland Mexico (purple), series Incarnatae sensu Fishbein (pink), Sonoran Desert (blue), and outgroup (black).
CONCLUSIONS
Hyb-Seq, the combined target enrichment and genome skimming approach presented here, efficiently generates copious data from both the low-copy nuclear genome and high-copy elements (e.g., organellar genomes) appropriate for phylogenomic analyses in plants. With a small investment to generate a genome and transcriptome for an exemplar or the utilization of quickly growing resources from the many publicly available genome and transcriptome projects, a probe set can be designed that will target conserved regions that are phylogenetically informative across plant genera or families. Because this approach recovers sequences that are hundreds of base pairs in length from hundreds to thousands of loci, even with modest levels of variation the data are appropriate for addressing questions at low taxonomic levels. Furthermore, sequences flanking the conserved target regions will generally evolve more rapidly, providing additional potentially informative variation.
The Hyb-Seq protocol based on taxon-specific genome and transcriptome data has advantages over alternative approaches, such as transcriptome sequencing or genome reduction via restriction digest. Transcriptome sequencing results in thousands of orthologous nuclear loci, but requires living, flash frozen, or specially preserved tissue for RNA extraction, is subject to large amounts of missing loci across samples, and does not as effectively sample rapidly evolving noncoding regions. In contrast, target capture and genome skimming can use small amounts of relatively degraded DNA, such as extractions from herbarium specimens (Cronn et al., 2012; Straub et al., 2012), and consistently yield intron and 5′ and 3′ untranslated region sequence. Genome reduction methods utilizing restriction digests (e.g., RAD-Seq, genotyping-by-sequencing; Davey et al., 2011) also produce thousands of loci, and have been effective in resolving phylogenetic relationships and patterns of introgression (e.g., Eaton and Ree, 2013). However, the effectiveness of these approaches with poor quality or degraded DNA has not been demonstrated, and the anonymous nature of these loci makes it more challenging to determine orthology. Most importantly, the data obtained (SNPs or 30–200-bp sequences) are not appropriate for applying phylogenetic approaches that estimate species trees from a large number of gene trees. Focusing on orthologous targets through Hyb-Seq also reduces the amount of missing data relative to both transcriptome and RAD-Seq studies. Until the sequencing of whole genomes for every species of interest becomes practical and affordable, the protocol presented here is poised to become the standard for quickly and efficiently producing genome-scale data sets to best advance our understanding of the evolutionary history of plants.
Supplementary Material
Appendix
Appendix 1.
Voucher information for species of Asclepias and related genera used in this study.
| Species | Voucher specimen [Herbarium] | Collection locality | GPS coordinatesa |
| Asclepias cryptoceras S. Watson | Weitemier 12-23 [OSC] | Grant Co., Oregon, USA | 44.47970, −119.57758 |
| A. engelmanniana Woodson | Lynch 11224 [LSUS] | Barber Co., Kansas, USA | 37.3, −98.7 |
| A. eriocarpa Benth. | Lynch 10923 [LSUS] | Lassen Co., California, USA | 41.09, −121.30 |
| A. flava (Kuntze) Lillo non N. E. Br. | Zuloaga & Morrone 7069 [OKLA] | Dist. Jujuy, Argentina | −24, −63.35 |
| A. humistrata Walter | Fishbein 5596 [OKLA] | Polk Co., Florida, USA | 27.761, −81.465 |
| A. involucrata Englem. ex Torr. | Lynch 12050 [LSUS] | Apache Co., Arizona, USA | 36.7, −109.7 |
| A. masonii Woodson | Fishbein 3101 [OKLA] | Mpio. Comondu, Baja California Sur, Mexico | 24.63, −112.14 |
| A. nyctaginifolia A. Gray | Fishbein 2445 [ARIZ] | Pima Co., Arizona, USA | 31.80, −110.81 |
| A. scheryi Woodson | Fishbein 5137 [OKLA] | Mpio. Cuautitlán, Jalisco, Mexico | 19.561, −114.203 |
| A. tomentosa Elliott | Fishbein 5608 [MISSA] | Franklin Co., Florida, USA | 29.916, −84.369 |
| Calotropis procera (Aiton) W. T. Aiton | Fishbein 5427 [OKLA] | Cultivated | |
| Matelea cynanchoides (Engelm. & A. Gray) Woodson | Rein 106 [OKLA] | Angelina Co., Texas, USA | 31.07995, −94.27735 |
GPS coordinates reported to the accuracy recorded or based on coarse geo-referencing based on the collection locality.
LITERATURE CITED
- Bolger A. M., Lohse M., Usadel B. 2014. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics (Oxford, England) 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman M. A., Chang J., Weisman D., Kesseli R. V., Burke J. M. 2007. Universal markers for comparative mapping and phylogenetic analysis in the Asteraceae (Compositae). Theoretical and Applied Genetics 115: 747–755 [DOI] [PubMed] [Google Scholar]
- Cronn R., Knaus B. J., Liston A., Maughan P. J., Parks M., Syring J. V., Udall J. 2012. Targeted enrichment strategies for next-generation plant biology. American Journal of Botany 99: 291–311 [DOI] [PubMed] [Google Scholar]
- Davey J. W., Hohenlohe P. A., Etter P. D., Boone J. Q., Catchen J. M., Blaxter M. L. 2011. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nature Reviews. Genetics 12: 499–510 [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15 [Google Scholar]
- Duarte J. M., Wall P. K., Edger P. P., Landherr L. L., Ma H., Pires J. C., Leebens-Mack J., et al. 2010. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evolutionary Biology 10: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton D. A. R., Ree R. H. 2013. Inferring phylogeny and introgression using RADseq data: An example from flowering plants (Pedicularis: Orobanchaceae). Systematic Biology 62: 689–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faircloth B. C. 2014. phyluce Phylogenetic estimation from ultraconserved elements. 10.6079/J9PHYL. GitHub repository https://github.com/faircloth-lab/phyluce [accessed 15 July 2014].
- Faircloth B. C., McCormack J. E., Crawford N. G., Harvey M. G., Brumfield R. T., Glenn T. C. 2012. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Systematic Biology 61: 717–726 [DOI] [PubMed] [Google Scholar]
- Fishbein M., Chuba D., Ellison C., Mason-Gamer R. J., Lynch S. P. 2011. Phylogenetic relationships of Asclepias (Apocynaceae) inferred from non-coding chloroplast DNA sequences. Systematic Botany 36: 1008–1023 [Google Scholar]
- Gongora-Castillo E., Childs K. L., Fedewa G., Hamilton J. P., Liscombe D. K., Magallanes-Lundback M., Mandadi K. K., et al. 2012. Development of transcriptomic resources for interrogating the biosynthesis of monoterpene indole alkaloids in medicinal plant species. PLoS ONE 7: e52506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A. 2010. FASTX-Toolkit. Website http://hannonlab.cshl.edu/fastx_toolkit/ [accessed 15 May 2014].
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wickett N. J., Ayyampalayam S., Chanderbali A. S., Landherr L., Ralph P. E., Tomsho L. P., et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Katoh K., Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics 9: 286–298 [DOI] [PubMed] [Google Scholar]
- Kent W. J. 2002. BLAT—the BLAST-Like Alignment Tool. Genome Research 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus B. 2012. Short read toolbox. Website http://brianknaus.com/software/srtoolbox/ [accessed 15 May 2014].
- Kubatko L., Degnan J. 2007. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Systematic Biology 56: 17–24 [DOI] [PubMed] [Google Scholar]
- Lemmon E. M., Lemmon A. R. 2013. High-throughput genomic data in systematics and phylogenetics. Annual Review of Ecology Evolution and Systematics 44: 99–121 [Google Scholar]
- Li C., Hofreiter M., Straube N., Corrigan S., Naylor G. J. 2013. Capturing protein-coding genes across highly divergent species. BioTechniques 54: 321–326 [DOI] [PubMed] [Google Scholar]
- Li W., Godzik A. 2006. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics (Oxford, England) 22: 1658–1659 [DOI] [PubMed] [Google Scholar]
- Liu L., Yu L., Edwards S. V. 2010. A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evolutionary Biology 10: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel J. R., Dikow R. B., Funk V. A., Masalia R. R., Staton S. E., Kozik A., Michelmore R. W., et al. 2014. A target enrichment method for gathering phylogenetic information from hundreds of loci: An example from the Compositae. Applications in Plant Sciences 2(2):1300085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack J. E., Harvey M. G., Faircloth B. C., Crawford N. G., Glenn T. C., Brumfield R. T. 2013. A phylogeny of birds based on over 1,500 loci collected by target enrichment and high-throughput sequencing. PLoS ONE 8: e54848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata J., Bienzle D., Brandle J. E., Sensen C. W., De Luca V. 2006. Expressed sequence tags from Madagascar periwinkle (Catharanthus roseus). FEBS Letters 580: 4501–4507 [DOI] [PubMed] [Google Scholar]
- Nylander J. A. A. 2011. Catfasta2pyml.pl. Website http://www.abc.se/∼nylander/catfasta2phyml/ [accessed 15 May 2014].
- Parks M., Cronn R., Liston A. 2012. Separating the wheat from the chaff: Mitigating the effects of noise in a plastome phylogenomic data set from Pinus L. (Pinaceae). BMC Evolutionary Biology 12: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratan A. 2009. Assembly algorithms for next-generation sequence data. Ph.D. dissertation, The Pennsylvania State University, University Park, Pennsylvania, USA.
- Reneker J., Lyons E., Conant G. C., Pires J. C., Freeling M., Shyu C.-R., Korkin D. 2012. Long identical multispecies elements in plant and animal genomes. Proceedings of the National Academy of Sciences, USA 109: E1183–E1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salichos L., Rokas A. 2013. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497: 327–331 [DOI] [PubMed] [Google Scholar]
- Shaw T. I., Ruan Z., Glenn T. C., Liu L. 2013. STRAW: Species TRee Analysis Web server. Nucleic Acids Research 41: W238–W241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J. T., Wong K., Jackman S. D., Schein J. E., Jones S. J. M., Birol İ. 2009. ABySS: A parallel assembler for short read sequence data. Genome Research 19: 1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics (Oxford, England) 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Straub S. C. K., Fishbein M., Livshultz T., Foster Z., Parks M., Weitemier K., Cronn R. C., Liston A. 2011. Building a model: Developing genomic resources for common milkweed (Asclepias syriaca) with low coverage genome sequencing. BMC Genomics 12: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub S. C. K., Parks M., Weitemier K., Fishbein M., Cronn R. C., Liston A. 2012. Navigating the tip of the genomic iceberg: Next-generation sequencing for plant systematics. American Journal of Botany 99: 349–364 [DOI] [PubMed] [Google Scholar]
- Stull G. W., Moore M. J., Mandala V. S., Douglas N. A., Kates H.-R., Qi X., Brockington S. F., et al. 2013. A targeted enrichment strategy for massively parallel sequencing of angiosperm plastid genomes. Applications in Plant Sciences 1(2):1200497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. A., Govindarajulu R., Liston A., Ashman T.-L. 2013. Targeted sequence capture provides insight into genome structure and genetics of male sterility in a gynodioecious diploid strawberry, Fragaria vesca ssp. bracteata (Rosaceae). G3·Genes|Genomes|Genetics 3: 1341–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Mueller L. A., Crouzillat D., Petiard V., Tanksley S. D. 2006. Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: A test case in the euasterid plant clade. Genetics 174: 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer E. A., Wen J. 2013. Using nuclear gene data for plant phylogenetics: Progress and prospects. Molecular Phylogenetics and Evolution 66: 539–550 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.