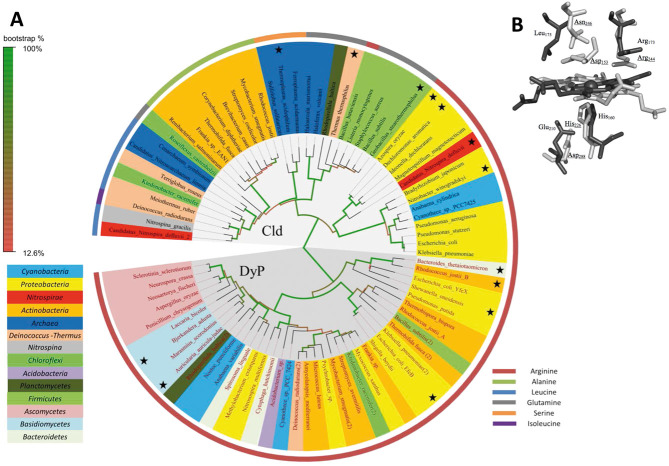
Figure 1.
Phylogeny and active site architecture of clorite dismutase (Cld) and dye-decolorizing peroxidase (DyP). (A) Maximum likelihood tree based on amino acid sequences of Clds, Cld-like proteins and DyPs. Sequences marked with an asterisk represent proteins of known X-ray structure. Colors highlighting the species name indicate the affiliations of the respective organisms to bacterial and archaeal phylae based on 16S rRNA phylogeny. Color of the stripe, bordering the figure, define the residues at the position which is homologous to Arg173 of NdCld, in the respective proteins. This residue is fully conserved in functional (i.e. chlorite degrading) Clds. The circular tree was drawn using FigTree v1.4 (http://tree.bio.ed.ac.uk/software/figtree/). (B) Overlay of active site of Cld and DyP. Cld from “Candidatus Nitrospira defluvii” (NdCld, PDB: 3NN1) is depicted in dark gray, and DypB from Rhodococcus jostii (PDB: 3QNS) in light gray; Amino acid numbering follows NdCld and DypB from Rhodococcus jostii (underlined). Figure was generated using PyMOL (http://www.pymol.org/).