Figure 2.
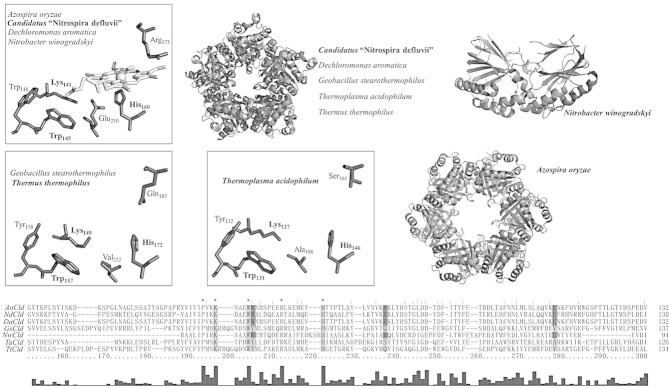
Crystal structures and sequence alignment of Clds and Cld-like proteins. Oligomeric structures and active site residues of Clds from Azospira oryzae (2VXH), Candidatus Nitrospira defluvii (3NN1), Dechloromonas aromatica (3Q08, 3Q09), and Nitrobacter winogradskyi (3QPI). Oligomeric structures and heme cavity residues in Cld-like proteins from Geobacillus stearothermophilus (1T0T), Thermus thermophilus (1VDH) and Thermoplasma acidophilum (3DTZ). Note that these X-ray structures do not contain the prosthetic group. Cld structures from species written in bold are depicted. Residues in the sequence alignment of the C-terminal domains, which are depicted in the structures, are highlighted in light gray; depicted residues unique for Clds with chlorite decomposition activity are highlighted in dark gray. Bar graph shows conserved amino acids. Figures were generated using PyMOL (http://www.pymol.org/).
