Figure 6. Cdc48p-dependent in vitro de-ubiquitination by Otu1p.
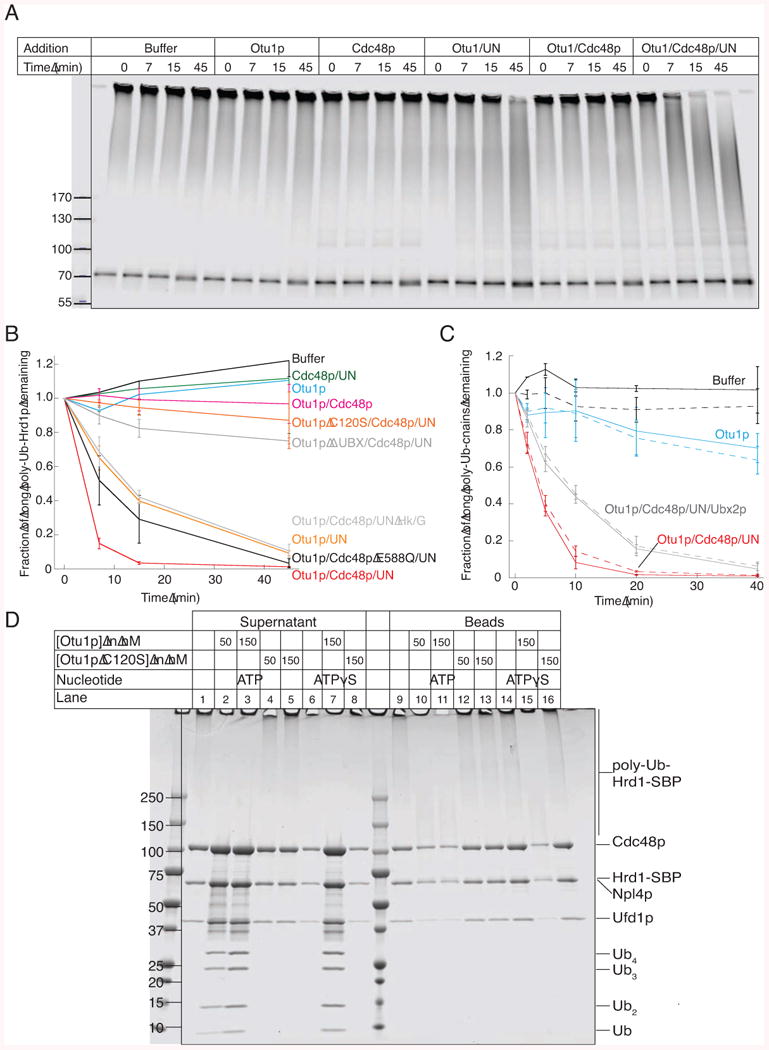
(A) Bead-immobilized fluorescently labeled Hrd1p-SBP was incubated with the ubiquitination machinery. After washing, Hrd1p was eluted from the beads with biotin and incubated with the indicated components (Ufd1/Npl4p; UN) for different time periods in the presence of ATP. Hrd1p was in a 30-fold excess over Otu1p, whereas all other components were about equimolar to Hrd1p. The samples were analyzed by SDS-PAGE and fluorescence-scanning.
(B) Quantification of experiments performed as in (A). The disappearance of the longest ubiquitin chains was quantified under different conditions (means and standard deviation of three experiments). ATP was depleted with hexokinase/glucose (Hk/G). Where indicated, an ATPase-defective Cdc48p mutant (Cdc48p E588Q) or an Otu1p mutant lacking the Ubx domain were used.
(C) Bead-immobilized complexes of Hrd1p-SBP and CPY*, labeled with different fluorescent dyes, were treated as in (A). The de-ubiquitination of modified Hrd1p and CPY* was followed in parallel (solid and broken lines, respectively). Shown are the means and standard deviations of three experiments.
(D) Bead-immobilized ubiquitinated Hrd1-SBP was incubated with Cdc48p complex in the presence of ATPγS. After washing, the beads were incubated for 1 h with the indicated components. Supernatants and beads were analyzed by SDS-PAGE and Coomassie staining.
See also Figure S6.
