Abstract
The gut microbiota is involved in metabolic and immune disorders associated with obesity and type 2 diabetes. We previously demonstrated that prebiotic treatment may significantly improve host health by modulating bacterial species related to the improvement of gut endocrine, barrier and immune functions. An analysis of the gut metagenome is needed to determine which bacterial functions and taxa are responsible for beneficial microbiota–host interactions upon nutritional intervention. We subjected mice to prebiotic (Pre) treatment under physiological (control diet: CT) and pathological conditions (high-fat diet: HFD) for 8 weeks and investigated the production of intestinal antimicrobial peptides and the gut microbiome. HFD feeding significantly decreased the expression of regenerating islet-derived 3-gamma (Reg3g) and phospholipase A2 group-II (PLA2g2) in the jejunum. Prebiotic treatment increased Reg3g expression (by ∼50-fold) and improved intestinal homeostasis as suggested by the increase in the expression of intectin, a key protein involved in intestinal epithelial cell turnover. Deep metagenomic sequencing analysis revealed that HFD and prebiotic treatment significantly affected the gut microbiome at different taxonomic levels. Functional analyses based on the occurrence of clusters of orthologous groups (COGs) of proteins also revealed distinct profiles for the HFD, Pre, HFD-Pre and CT groups. Finally, the gut microbiota modulations induced by prebiotics counteracted HFD-induced inflammation and related metabolic disorders. Thus, we identified novel putative taxa and metabolic functions that may contribute to the development of or protection against the metabolic alterations observed during HFD feeding and HFD-Pre feeding.
Keywords: metagenomic, gut microbiota, type 2 diabetes, antimicrobial peptides, Reg3g, prebiotics
Introduction
Obesity and related metabolic disorders are associated with low-grade inflammation, which contributes to the onset of these diseases (Olefsky and Glass, 2010). The gut microbiota influences whole-body metabolism by affecting energy balance and metabolic inflammation associated with obesity and related disorders (Tremaroli and Backhed, 2012; Everard and Cani, 2013). We and others have previously demonstrated that high-fat diet (HFD) feeding changes gut microbiota composition (Cani et al., 2007a, 2007b; Turnbaugh et al., 2008; Hildebrandt et al., 2009; Everard et al., 2013) and identified serum lipopolysaccharides (LPS) (that is, metabolic endotoxemia) as a novel factor linking gut microbiota with the onset of inflammation and insulin resistance associated with obesity (Cani et al., 2007a, 2009). We have contributed to the demonstration that obesity and type 2 diabetes are associated with increased gut permeability, thereby inducing metabolic endotoxemia and associated inflammation (Cani et al., 2009). Compelling evidence suggests that oral supplementation with selectively fermented oligosaccharides (that is, prebiotics) improves these metabolic disorders via several mechanisms (Guarner, 2007; Cani et al., 2009; Muccioli et al., 2010; Everard et al., 2011, 2013). For example, we discovered that feeding genetic or diet-induced obese mice with prebiotics increased the abundance of Akkermansia muciniphila (A. muciniphila) by ∼100-fold, which was correlated with an improved metabolic status (Everard et al., 2011, 2013). Recently, we have uncovered novel mechanisms of interaction between this bacterium and the host. We demonstrated that A. muciniphila treatment reversed HFD-induced metabolic disorders (that is, reduced fat-mass gain, metabolic endotoxemia, adipose tissue inflammation and insulin resistance) by mechanisms associated with the restoration of adequate intestinal mucus production by goblet cells, which consequently improve barrier function (Everard et al., 2013). The mucus barrier produced by goblet cells is reinforced by antimicrobial peptides associated with innate immunity and produced by Paneth cells (for example, α-defensins, lysozyme C, phospholipases and C-type lectins, namely regenerating islet-derived 3-gamma, Reg3g) or by enterocytes (Reg3g) (Hooper and Macpherson, 2010; Bevins and Salzman, 2011; Pott and Hornef, 2012). Importantly, we determined that HFD feeding decreases intestinal Reg3g expression, whereas oral supplementation with A. muciniphila counteracted this effect (Everard et al., 2013). These immune factors constitute key factors involved in host–gut microbiota interactions. Indeed, through these immune factors, the host controls its interactions with the gut microbiota and thereby shapes its microbial communities (Pott and Hornef, 2012).
Complex rearrangements and constant intestinal epithelium renewal are also involved in intestine homeostasis without compromising epithelial barrier integrity (Vereecke et al., 2011). However, this process of cell shedding under homeostatic condition must be tightly regulated to preserve the integrity of the gut barrier. Prebiotic treatment improves gut barrier functions through several mechanisms (Cani et al., 2009; Muccioli et al., 2010). However, it is unknown whether this constant intestinal epithelium turnover is affected by prebiotics.
Although it is well established that diet-induced obesity is associated with changes in gut microbiota composition, few data are available regarding the impact of HFD feeding on metagenomic changes, and no study has investigated the intestinal host response (Turnbaugh et al., 2008, 2009; Hildebrandt et al., 2009). We have previously reported that prebiotic treatment changes the proportion of >100 taxa in genetic obese mice (Everard et al., 2011), but the impact of prebiotics on the gut metagenome under both physiological and diet-induced obese conditions remains unknown. Moreover, whether dietary interventions such as HFD or prebiotic supplementation affect the production of antimicrobial peptides has not been investigated.
Thus, this study aims (i) to elucidate the impact of HFD feeding or prebiotic treatment (under normal diet or an HFD) on the taxonomic profile and metabolic functions of the mouse gut microbiome and (ii) to investigate the influence of such dietary interventions on host antimicrobial peptide production. We used deep metagenomic sequencing analysis of caecal contents to demonstrate that both HFD and prebiotics independently affect the gut microbiome. We also linked gut microbial composition and functions with the production of specific host antimicrobial peptides.
Materials and methods
Mice
A set of 10-week-old C57BL/6J mice (40 mice, n=10 per group) (Charles River Laboratories, Brussels, Belgium) were housed in groups of 5 mice per cage, with free access to food and water. The mice were fed a control diet (CT) (A04, Villemoisson-sur-Orge, France), a CT supplemented with prebiotics (oligofructose) (Orafti, Tienen, Belgium) (0.3 g per mouse per day) added in tap water (CT-Pre), an HFD (60% fat and 20% carbohydrates (kcal per 100 g), D12492, Research Diet, New Brunswick, NJ, USA), or an HFD supplemented with oligofructose (0.3 g per mouse per day) added in tap water (HFD-Pre). The treatment continued for 8 weeks. This set of mice was previously metabolically characterised in Everard et al. (2013).
All mice experiments were approved by and performed in accordance with the guidelines of the local ethics committee. Housing conditions were specified by the Belgian Law of May 29 2013 regarding the protection of laboratory animals (agreement number LA1230314).
Tissue sampling
The animals were anaesthetised with isoflurane (Forene; Abbott, Queenborough, Kent, England) before exsanguination and tissue sampling; the mice were then killed by cervical dislocation. Intestinal segments (jejunum and colon) and caecal contents were collected at death, immersed in liquid nitrogen, and stored at –80 °C until further analysis.
Insulin resistance index
Plasma insulin concentration was determined using an ELISA kit (Mercodia, Upssala, Sweden) according to the manufacturer's instructions. Insulin resistance index was determined by multiplying the area under the curve (0 and 15 min) of both blood glucose and plasma insulin obtained following an oral glucose load (2 g of glucose per kg of body weight) performed after 6 weeks of dietary treatment. Food was removed 2 h after the onset of the daylight cycle and mice were treated after 6-h fasting period as previously described (Everard et al., 2011).
Plasma leptin measurement
Leptin levels were measured in cava vein plasma using ELISA according to the manufacturer's instructions (Mouse ELISA leptin, EZML-82K; Merck Millipore, Darmstadt, Germany).
SCFA caecal content
Short chain fatty acids (SCFA) levels in caecal content were analysed using gas chromatography coupled to a mass spectrometer as previously described (Wichmann et al., 2013).
RNA preparation and real-time qPCR analysis
Total RNA was prepared from tissues using TriPure reagent (Roche Diagnostics, Mannheim, Germany). Quantification and integrity analysis of total RNA were performed by analysing 1 μl of each sample in an Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit; Agilent, Waghaeusel-Wiesental, Belgium). cDNA was prepared by reverse transcription of 1 μg total RNA using a Reverse Transcription System kit (Promega, Leiden, The Netherlands). Real-time PCR was performed with the StepOnePlus real-time PCR system and software (Applied Biosystems, Den Ijssel, The Netherlands) using Mesa Fast qPCR (Eurogentec, Seraing, Belgium) for detection according to the manufacturer's instructions. RPL19 RNA was chosen as the housekeeping gene. All samples were performed in duplicate in a single 96-well reaction plate, and data were analysed according to the 2−ΔCT method. The identity and purity of the amplified product was assessed by melting curve analysis at the end of amplification. The primer sequences for the targeted mouse genes are presented in Supplementary Table S4.
DNA isolation from mouse caecal samples
Metagenomic DNA was extracted from the caecal content using a QIAamp-DNA stool mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions and the adapted procedure described in Dewulf et al. (2013). Based on the quantity and the quality of the DNA extracted, samples were selected to perform the sequencing.
Sequencing
Metagenomic DNA fragment libraries that were prepared according to Illumina instructions were indexed using 6-base sequences. The libraries were sequenced from a single end for 100+7 cycles on an Illumina HiSeq 2000 using the TruSeq SBS v3 kit (Illumina, San Diego, CA, USA). The PhiX reference was spiked in relevant channels of the flow cell to determine whether the error rate was within Illumina specifications (⩾80% of the reads with a Q30 error rate of below 1.5%). Base-calling was performed with HiSeq Control software 1.5.15.1, RTA 1.13.48.0 and CASAVA 1.8.2 (Illumina). Sequence reads were first filtered using the default Illumina quality criteria.
Any base at the 5'-terminus was removed if its quality score was ⩽Q10. The reads were further filtered by the average quality score Q30 over a sliding 20-base window and a minimum sequence length of 80 bases using the Mothur (1.26) (Schloss et al., 2009) command trim.seqs. Sequences were deposited in MG-RAST under the project ID 6153.
Sequence analysis
Taxonomic analysis
Sequences were compared with the Greengenes reference 16S rRNA gene database pre-clustered at 97% identity (Greengenes file gg_97_otus_4feb2011.fasta) (McDonald et al., 2012) using BLASTN (-evalue 1e-030 -perc_identity 97 -max_target_seqs 1) (Altschul et al., 1990). Minimum query alignment coverage was set to 90%.
Functional analysis
We used a stand-alone version of DECONSEQ (Schmieder and Edwards, 2011) with ⩾90% coverage and ⩾94% identity to sequentially extract sequences matching mouse (em_rel_std_mus), viral (em_rel_std_vrl) and fungal (em_rel_std_fun) databases. The remaining sequences were considered to be mainly derived from bacterial DNA. From the sequences assigned to bacteria, open reading frames were identified using FragGeneScan (Rho et al., 2010) (parameters –complete 0 –train illumina_5). To functionally annotate protein sequences, we used the CAMERA (Sun et al., 2011) or WebMGA (Wu et al., 2011) function prediction workflow and the NCBI COG database for prokaryotic proteins (Tatusov et al., 2003; Klimke et al., 2009). All hits below the default RPSBLAST e-value of 1e-03 were reported.
Clustering of bacterial communities
To compare bacterial communities, we constructed a Bray–Curtis similarity matrix based on the square-root transformed relative abundances of operational taxonomic unit or clusters of orthologous group (COG) functions. Principal coordinate analysis of Bray–Curtis similarities was performed in PRIMER-E (Primer-E Ltd, Plymouth, UK). Ecological indices were calculated from operational taxonomic unit relative abundances in PRIMER-E.
Statistical analysis
Data are expressed as means±s.e.m. unless otherwise indicated. Differences between two groups were assessed using the unpaired two-tailed Student's t-test, Mann–Whitney U test and Permanova (Primer-E Ltd). Data sets that involved more than two groups were assessed by ANOVA followed by Newman–Keuls post hoc tests. In the figures, data with different superscript letters are significantly different at P<0.05, according to post hoc ANOVA statistical analyses. Data were analysed using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA, USA). The results were considered as statistically significant at P<0.05.
Results
HFD feeding and prebiotic treatment profoundly affect the expression of intestinal antimicrobial peptides
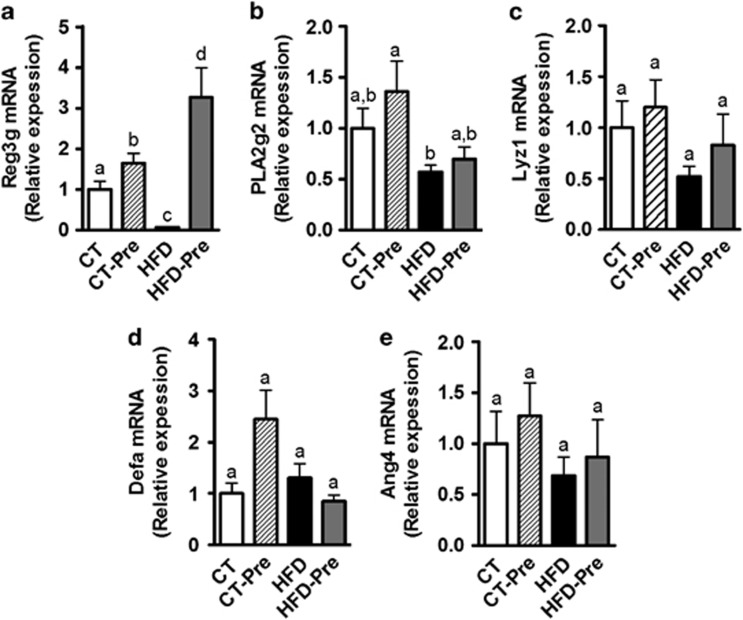
Antimicrobial peptides produced by the host have an important role in maintaining gut microbiota homeostasis and physical segregation of commensal microorganisms from host tissue. These peptides constitute an attractive mechanism for gut ecosystem modulation upon HFD feeding or upon prebiotic treatment. Consistent with this hypothesis, we observed that HFD feeding affected antimicrobial peptide production in the small intestine as well as in the colon. In the small intestine, the expression of Reg3g (RegIIIγ) was decreased upon HFD feeding (Figure 1a), whereas phospholipase A2 group II (Pla2g2), Lysozyme C (Lyz1) and angiogenin 4 (Ang4) mRNA expression tended to be reduced by the HFD (Figures 1b, c and e), and α-defensin (Defa) expression was similar to that in the control group (Figure 1d). Prebiotic treatment increased Reg3g expression in the small intestine upon control or HFD feeding, whereas other antimicrobial peptides were not modified by the treatment (Figure 1). We previously demonstrated that HFD feeding decreases Reg3g expression in the colon (Everard et al., 2013). Here, we found that prebiotic treatment increased Reg3g expression in the colon by ∼6-fold (HFD 1±0.25 vs HFD-Pre 5.88±1.42, P=0.0049).
Figure 1.
HFD feeding and prebiotic treatment affect antimicrobial peptides in the intestine. Antibacterial peptide mRNA expression: (a) Regenerating islet-derived 3-gamma (RegIIIγ, encoded by Reg3g); (b) Phospholipase A2 group II (encoded by Pla2g2); (c) Lysozyme C (encoded by Lyz1); (d) α-defensins (encoded by Defa); and (e) Angiogenin 4 (encoded by Ang4) measured in the jejunum of control diet-fed mice (CT) (n=9), CT diet-fed mice treated with prebiotics (CT-Pre) (n=10), HFD-fed mice (HFD) (n=10) and HFD-fed mice treated with prebiotics (HFD-Pre) (n=10). Data are means±s.e.m. Data with different superscript letters are significantly different (P<0.05) according to a post hoc ANOVA one-way statistical analysis.
Therefore, the expression of transcripts for antimicrobial peptides is affected by the HFD vs prebiotic treatment. Since these peptides are key factors involved in shaping gut microbiota (Gallo and Hooper, 2012), we also assessed caecal bacterial communities using taxonomic and functional metagenomic approaches.
Prebiotic treatment increases the expression of intectin, a key protein involved in intestinal epithelial cell turnover
The gut mucosa is subjected to a constant and rapid cellular turnover essential for maximal nutrient absorption, adaptation to changes in diet and repair of mucosal injury. It is commonly accepted that rapid cell renewal coincides with the apical exfoliation of enterocytes without necessarily compromising the gut barrier integrity or even more reinforcing the barrier function (Cliffe et al., 2005; Vereecke et al., 2011). The small intestine-specific glycosylphosphatidylinositol-anchored protein, intectin, has been shown to be distinctly localised at the villus tips of the intestinal mucosa (Kitazawa et al., 2004). Importantly, intectin has been proposed to be involved in the rapid turnover of intestinal mucosa (Kitazawa et al., 2004). Here, we found that prebiotic treatment significantly increases the mRNA expression of intectin by 3- to 5-fold under CT or HFD, respectively. These findings suggest that prebiotic feeding increases epithelial cell turnover (Figure 2), which could constitute a new mechanism contributing to reinforce the intestinal barrier induced by prebiotics (Cliffe et al., 2005; Vereecke et al., 2011).
Figure 2.
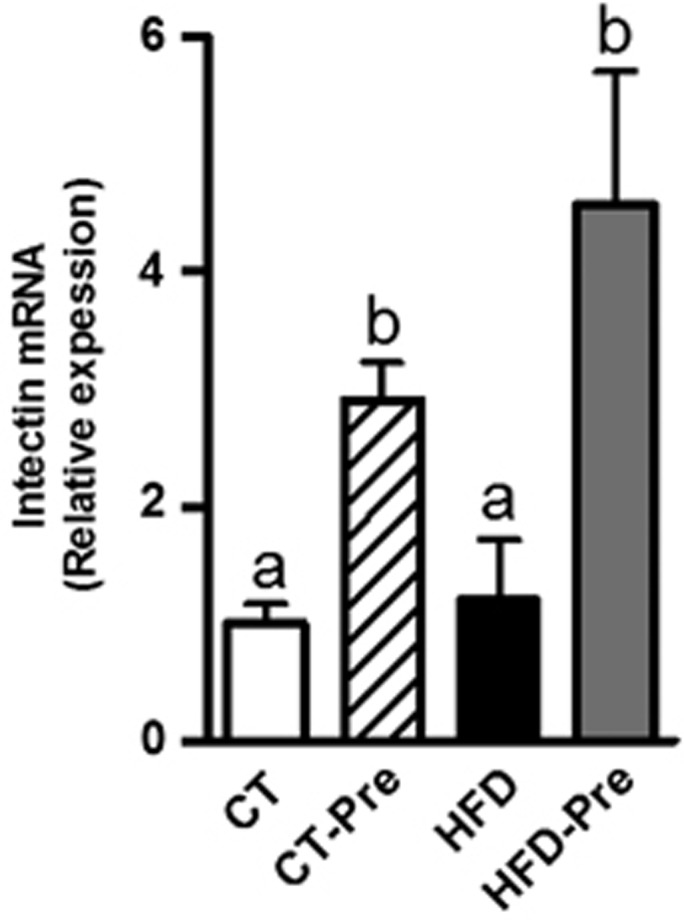
Prebiotic treatment increases colon intectin expression, a protein involved in the turnover of intestinal mucosa. Intectin mRNA expression measured in the colon of control diet-fed mice (CT) (n=9), CT diet-fed mice treated with prebiotics (CT-Pre) (n=10), HFD-fed mice (HFD) (n=10) and HFD-fed mice treated with prebiotics (HFD-Pre) (n=10). Data are means±s.e.m. Data with different superscript letters are significantly different (P<0.05) according to a post hoc ANOVA one-way statistical analysis.
HFD feeding and prebiotic treatment profoundly affect the taxonomic composition of the gut microbiome
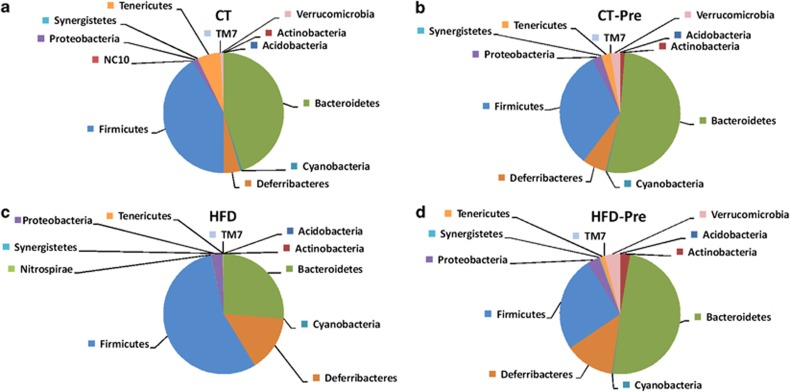
In accordance with previous studies (Everard et al., 2011), the mouse microbiome was greatly dominated by the phyla Firmicutes and Bacteroidetes (Figure 3). HFD treatment profoundly affected the caecal Firmicutes/Bacteroidetes ratio (Figures 3a and c; Supplementary Table S1) as well as the abundances of other phyla (Figure 3; Supplementary Table S1). The most important changes at the phylum level included a decrease in Tenericutes, Cyanobacteria and Verrucomicrobia (Figure 3c). Interestingly, under the HFD, prebiotic treatment decreased the Firmicutes/Bacteroidetes ratio as well as the proportion of Tenericutes, Cyanobacteria and Verrucomicrobia (Figure 3d; Supplementary Table S1).
Figure 3.
HFD feeding and prebiotic treatment affect the proportions of different phyla. The composition of abundant bacterial phyla identified in the gut microbiota of (a) control diet-fed mice (CT) (n=9), (b) CT diet-fed mice treated with prebiotics (CT-Pre) (n=9), (c) HFD-fed mice (HFD) (n=7) and (d) HFD-fed mice treated with prebiotics (HFD-Pre) (n=10). Undetected phyla are not represented on the pie chart. The significant changes in specific phyla are shown in Supplementary Table S1.
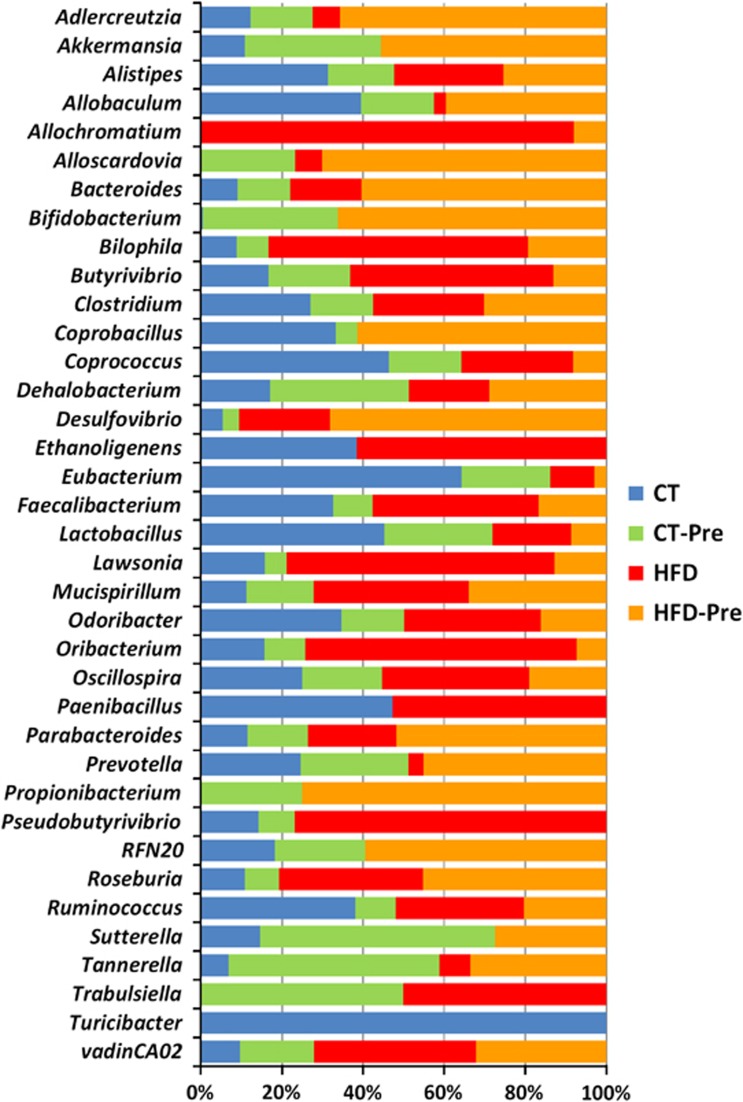
A total of 20 genera were significantly affected by the HFD compared with the CT, 8 of which belonged to the phylum Firmicutes. Under the HFD, we observed a global increase in Firmicutes, and some of its genera (Butyrivibrio, Oribacterium and Roseburia) followed that trend, whereas others (Allobaculum, Coprococcus, Eubacterium, Lactobacillus and Turicibacter) decreased drastically (Figure 4; Supplementary Table S2). Interestingly, Akkermansia, Bifidobacterium, Sutterella and Turicibacter were not detectable under HFD treatment alone but were detected (except for Turicibacter) upon prebiotic treatment in HFD-fed mice (Figure 4; Supplementary Table S2). The genera Allochromatium and Trabulsiella were present in HFD-induced obese mice and absent in control mice, whereas the genera Scardovia and Propionibacterium were identified only in prebiotic-treated (control and HFD) mice (Figure 4; Supplementary Table S2).
Figure 4.
HFD feeding and prebiotic treatment affect the proportions of different genera. The composition of the bacterial genera significantly affected by the treatment and identified in the gut microbiota of control diet-fed mice (CT) (n=9), CT diet-fed mice treated with prebiotics (CT-Pre) (n=9), HFD-fed mice (HFD) (n=7) and HFD-fed mice treated with prebiotics (HFD-Pre) (n=10). Each column is set at 100% to illustrate the proportion of each genus among the different groups; the absence of any colour indicates that the genus was not detected in this group of mice. The statistically significant changes observed between different groups are shown in Supplementary Table S2.
Moreover, we observed that the relative abundance of Bacteroides, Bilophila, Butyrivibrio, Mucispirullum, Oribacterium, Parabacteroides, Roseburia, vadinCA02 (Synergistaceae) and LE30 (Desulfovibrionaceae) was all significantly increased during HFD treatment (Figure 4; Supplementary Table S2). Among these genera, the prebiotic treatment under HFD significantly decreased the proportion of Bilophila, Butyrivibrio, LE30 and Oribacterium (Figure 4; Supplementary Table S2).
Conversely, Allobacullum, Coprococcus, Eubacterium, Lactobacillus and Prevotella were significantly decreased by the HFD treatment, whereas Allobacullum and Prevotella were increased by the prebiotic treatment in association with the HFD (Figure 4; Supplementary Table S2). Two other genera (Paenibacillus and Ethanoligenes) were not affected by the HFD treatment but decreased below the detection limit following prebiotic treatment for both diets (HFD or CT diet) (Figure 4; Supplementary Table S2).
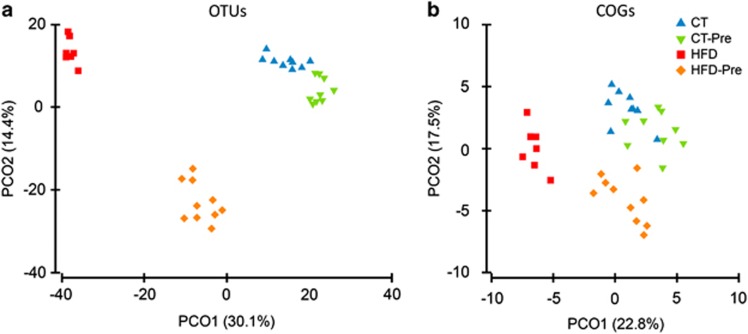
Multivariate analyses based on the operational taxonomic unit abundance revealed that the caecal microbiota clustered according to diet (Figure 5a). Statistical analysis confirmed a marked effect of the HFD on the caecal microbiota (Permanova t=3.56, P<0.002). Prebiotic intake had a lower overall impact on the microbiota than the HFD (Permanova t=1.946, P<0.002). The effect of prebiotic treatment was stronger (Permanova t=3.211, P<0.001) in HFD-fed mice than those fed the CT.
Figure 5.
PCoA of the gut bacterial communities and COG abundance. The analysis was based on the Bray–Curtis similarity matrix constructed using the square-root-transformed OTU (a) or COG abundances (b). The per cent of variation explained is given in brackets. OTU, operational taxonomic unit.
Functional analyses of the gut microbiome by COGs
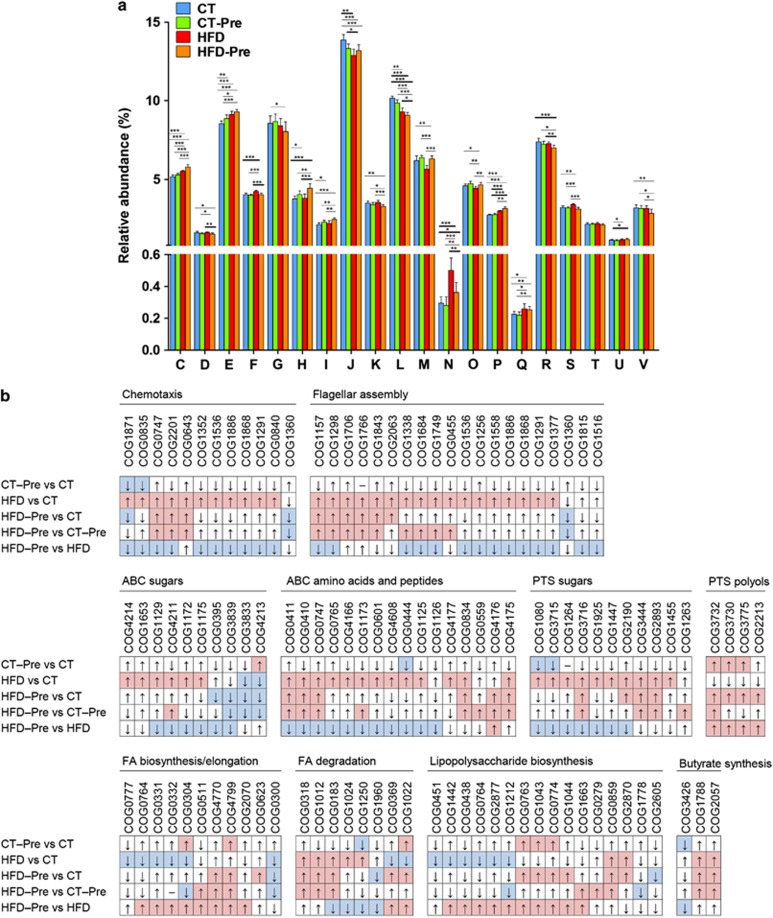
The gut microbiota assumes essential physiological functions in the host. Moreover, this huge potential functionality influences whole-body metabolism and is a key factor in the pathology of obesity. Therefore, to complement the taxonomic gut microbiota analyses, we performed functional analyses of the gut microbiome by assigning predicted open reading frames products to COGs. Globally, caecal microbiomes from mice fed different diets showed a similar distribution of the abundance of COG categories (classes) (Figure 6a). For 18 of 21 COG classes, we observed statistically significant differences between the CT and at least one of the three other diet groups. However, in most instances, these differences in COG proportions did not exceed 20% (Figure 6a).
Figure 6.
Orthologous groups of proteins (COGs) affected by the dietary treatments. (a) Occurrence of identified orthologous groups of proteins (COGs) according to COG functional categories: (A) RNA processing and modification are not shown on the figure since the abundance (%) was under 0.0025% in all four diet groups; (C) Energy production and conversion; (D) Cell-cycle control, cell division and chromosome partitioning; (E) Amino acid transport and metabolism; (F) Nucleotide transport and metabolism; (G) Carbohydrate transport and metabolism; (H) Coenzyme transport and metabolism; (I) Lipid transport and metabolism; (J) Translation, ribosomal structure and biogenesis; (K) Transcription; (L) Replication, recombination and repair; (M) Cell wall/membrane/envelope biogenesis; (N) Cell motility; (O) Posttranslational modification, protein turnover, chaperones; (P) Inorganic ion transport and metabolism; (Q) Secondary metabolites biosynthesis, transport and catabolism; (R) General function prediction only; (S) function unknown; (T), Signal transduction mechanisms; (U) Intracellular trafficking, secretion and vesicular transport; (V), defence mechanisms. Data are means±s.e.m. *P<0.05, **P<0.01, ***P<0.001 vs CT. (b) Changes in the occurrence of COGs according to the metabolic pathways affected following the different treatments. COGs with a median of ⩾4 (∼0.005% of the total number of COGs in normalised data sets) in at least one of the four diet groups were compared. Red and blue fields correspond to statistically significant increases and decreases in the COG relative abundances, respectively. The details of each COG number are shown in Supplementary Table S3.
Prebiotic treatment resulted in a statistically significant increase in the COG categories ‘Amino acid transport and metabolism', ‘Coenzyme transport and metabolism' and ‘Lipid transport and metabolism', whereas the COG categories ‘Translation, ribosomal structure and biogenesis' and ‘Replication, recombination and repair' had a lower proportion (Figure 6a; Supplementary Table S3). These COG classes shifted in the same direction under HFD and HFD-Pre treatments, but the changes in their abundance observed in the HFD-Pre vs CT comparisons had greater amplitude and larger statistical significance than those from the CT-Pre vs CT comparisons.
The HFD had a higher overall impact on the caecal bacterial functions than the prebiotic treatment. However, the effect of prebiotics was much stronger under the HFD than under the CT diet. In HFD-fed mice, the COG classes ‘Energy production and conversion', ‘Nucleotide transport and metabolism', ‘Inorganic ion transport and metabolism', ‘Amino acid transport and metabolism', ‘Cell Motility' and ‘Secondary metabolites biosynthesis, transport and catabolism' were enriched, whereas ‘Cell wall/membrane/envelope biogenesis' and ‘Posttranslational modification, protein turnover, chaperones' were depleted. The ‘Cell Motility' functions displayed the greatest shift induced by HFD, with a 69% increase relative to the CT group (Figure 6b; Supplementary Table S3). Most of the identified chemotaxis and flagellar assembly proteins were enriched by the HFD, and their abundance tended to decrease with the prebiotic-supplemented HFD (relatively to HFD) (Figure 6b; Supplementary Table S3).
The HFD was also correlated with an increased proportion of COGs corresponding to ABC transporters and sugar-specific phosphotransferase system (PTS) proteins, whereas the abundance of COGs related to sugar-alcohol-specific PTS proteins was reduced (Figure 6a; Supplementary Table S3). The opposite trends were observed upon addition of prebiotics to HFD-fed mice.
The proportion of COGs involved in fatty acid biosynthesis was lower under HFD, while COGs related to fatty acid degradation mainly increased in abundance. Again, addition of prebiotics to the HFD reversed these trends.
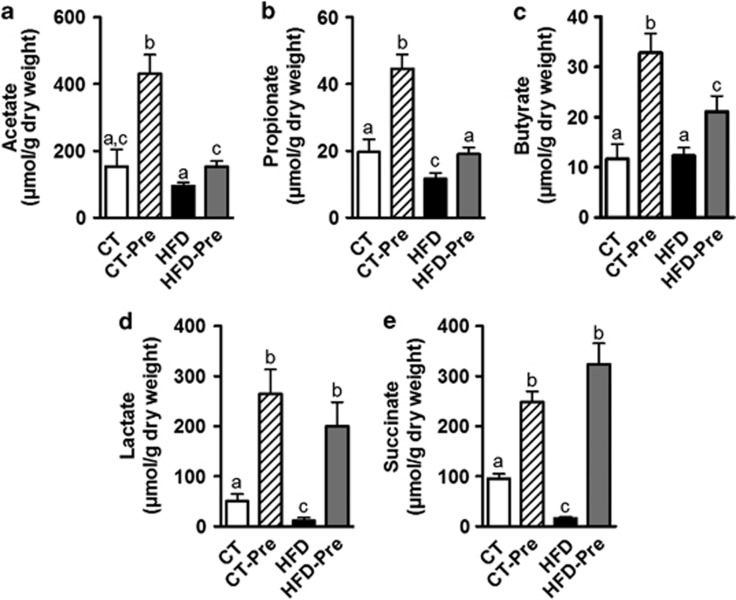
The proportion of COGs responsible for the synthesis of butyrate, which is an energy substrate for colonocytes, was affected by diet. Butyrate kinase, which corresponds to COG3426, was reduced in CT-Pre relative to the CT group (Supplementary Table S3). By contrast, two COGs potentially associated with butyrate production via a different pathway (COG1788 and COG2057, corresponding to acyl CoA:acetate/3-ketoacid CoA transferase subunits alpha and beta, respectively) significantly increased in abundance under HFD (Supplementary Table S3). Based on the abundance of these three COGs, butyrate production appeared to increase significantly under the HFD. Under both the CT and HFD diets, prebiotics apparently increased the butyrate synthesis potential via the CoA-transferase pathway, but these changes were not statistically significant (Figure 6b; Supplementary Table S3). Because this increase by HFD and the absence of prebiotic effects in butyrate synthesis potential is in contrast with recent studies (Daniel et al., 2013; Le Chatelier et al., 2013), we measured the SCFA levels in the caecal content. Importantly, we did not find any modification of acetate and butyrate levels in caecal content of HFD-fed mice compared with control mice whereas propionate, lactate and succinate were decreased by the HFD (Figure 7). However, prebiotic treatment increased the different SCFA (acetate, propionate, butyrate, lactate and succinate) under either CT diet or HFD (Figures 7a–e).
Figure 7.
Prebiotic treatment increases caecal short chain fatty acids content. (a) Actetate, (b) Propionate, (c) Butyrate, (d) Lactate and (e) Succinate concentrations (μmol g−1 of dry caecal content) measured in the caecal content of control diet-fed mice (CT) (n=9), CT diet-fed mice treated with prebiotics (CT-Pre) (n=10), HFD-fed mice (HFD) (n=10) and HFD-fed mice treated with prebiotics (HFD-Pre) (n=10). Data are means±s.e.m. Data with different superscript letters are significantly different (P<0.05) according to a post hoc ANOVA one-way statistical analysis.
Several COGs from the LPS biosynthetic pathway decreased in response to HFD and increased when the HFD-Pre microbiomes were compared with the HFD microbiomes (Supplementary Table S3).
HFD and prebiotic treatments affected the relative abundance of certain COGs in the same direction, while for the other COGs, they exerted opposite effects as indicated above (Supplementary Table S3). Identifying such effects of HFD and prebiotics could reveal novel mechanisms involved in the onset of obesity or in the improvement of the host metabolic status.
Principal coordinate analysis of the Bray–Curtis similarity matrix based on square-root-transformed COG proportions confirmed the clustering of the microbiota by the type of the diet revealed using the taxonomic approach (Figure 5b). PERMANOVA of Bray–Curtis similarities revealed a statistically significant effect of diet composition on overall caecal microbiota functions. The difference between the CT and CT-Pre groups (Permanova P<0.0004, t=1.585) was smaller than those found for any other pairwise comparisons between the four diet groups. The largest difference was observed when comparing mice treated with the HFD with those treated with prebiotics (CT-Pre) (Permanova t=4.402, P<0.0002). It seems that the functional potential of the caecal microbiome is less affected by the dietary treatments than is the taxonomic composition (Figure 5), and this is in accordance with the concept that the metagenomic carriage of metabolic pathways is more constant and more stable among individuals despite variation in community structure induced by environmental or dietary exposures (Human Microbiome Project Consortium, 2012). It should be noted, however, that a high proportion of metagenomic fragments were not assigned to any COGs. The role that this fraction of the metagenome may play in the bacterial (and host) metabolism remains unknown.
Caecal microbiota diversity
To assess the taxonomic and functional diversity of the caecal microbiota, we analysed the data sets normalised to 870 16S sequences and 82 417 COGs, respectively, as these figures corresponded to the sample with the smallest number of sequence reads. In the taxonomic analysis, the operational taxonomic unit richness and Shannon diversity index were lower in samples from the mice fed HFD relative to those from CT-fed mice, regardless of whether prebiotics were added (Table 1). Functional analysis revealed a slight but statistically significant increase in COG richness and diversity when HFD-Pre samples were compared with CT and CT-Pre groups. Therefore, taxonomic and functional measures do not necessarily follow the same direction of change. In response to specific conditions, an increase in functional diversity is possible despite a reduced taxonomic diversity. This may be due, for instance, to (i) enrichment in those bacterial species (or strains) that have a wider functional potential and (ii) an increase in the evenness of species (or strains) with different functional potentials.
Table 1. Ecological indices based on the relative abundance of OTUs and COG families.
|
Median (IQR) |
Change (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CT | CT-Pre | HFD | HFD-Pre | CT-Pre vs CT | HFD vs CT | HFD-Pre vs CT | HFD vs CT-Pre | HFD-Pre vs CT-Pre | HFD-Pre vs HFD | |
| 16S rDNA sequences normalised to 870 | ||||||||||
| Number of OTUs | 319 (308–339) | 291 (286–299) | 236 (231–247) | 259 (244–268) | ns | −26 (**) | −19 (***) | −19 (***) | −11 (**) | ns |
| Shannon diversity H' (loge) | 5.222 (5.053–5.309) | 5.051 (4.963–5.110) | 4.250 (4.200–4.470) | 4.485 (4.345–4.651) | ns | −19 (***) | −14 (***) | −16 (***) | −11 (***) | ns |
| Chao1 | 685 (550–755) | 606 (521–612) | 548 (465–596) | 555 (530–660) | ns | −20 (*) | ns | ns | ns | ns |
| COGs normalised to 82 417 | ||||||||||
| Number of COG families | 2188 (2177–2208) | 2194 (2190–2213) | 2190 (2176–2204) | 2244 (2227–2280) | ns | ns | 2.4 (***) | ns | 2.3 (**) | 2.4 (**) |
| Shannon diversity H' (loge) | 6.826 (6.807–6.827) | 6.827 (6.814–6.835) | 6.840 (6.834–6.846) | 6.876 (6.861–6.883) | ns | 0.34 (*) | 0.86 (***) | ns | 0.71 (***) | 0.52 (**) |
Abbreviations: COG, clusters of orthologous group; CT, control diet-fed mice; CT-Pre, CT diet-fed mice treated with prebiotics; HFD, HFD-fed mice; HFD-Pre, HFD-fed mice treated with prebiotics; IQR, interquartile range; OTU, operational taxonomic unit.
Mean of OTU and COG richness and the Shannon diversity index for the different treatment groups and percentage of changes.
Only significant changes (*P<0.05; **P<0.01; ***P<0.001 using Mann–Whitney test) are presented. ns, non-significant (P>0.05) changes.
Changes in gut microbiota taxa and functions counteracted HFD-induced inflammation, obesity and related metabolic disorders
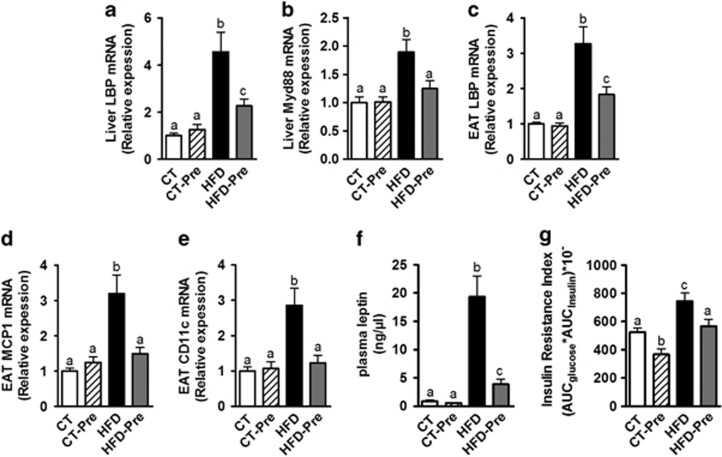
Importantly, gut microbiota modulations using prebiotics are associated with beneficial effects on obesity and associated metabolic disorders (Guarner, 2007; Cani et al., 2009; Muccioli et al., 2010; Everard et al., 2011, 2013). We have previously shown that the effects of prebiotics are related to an improvement of the gut barrier function resulting in the abolishment of metabolic endotoxemia (Cani et al., 2007b, 2009; Everard et al., 2013). Interestingly, previous reports have shown a strong association between the concentration of a circulating LPS-binding protein (LBP) and obesity-associated metabolic disorders (Ruiz et al., 2007; Sun et al., 2010; Moreno-Navarrete et al., 2012; Gonzalez-Quintela et al., 2013). It is worth noting that previous studies have shown an increase in plasma LBP during obesity or HFD-treated mice and a decrease during body weight loss (Fei and Zhao, 2013; Xiao et al., 2013). The LBP is mainly produced by the liver and enhances the sensitivity of cells to LPS (Hailman et al., 1994). Moreover, hepatic LBP production is a marker of LPS stimulation from the portal vein and is an indirect marker of portal endotoxin-induced hepatic LBP synthesis. Therefore, we measured the expression of LBP in the liver of our mice. We found that HFD induced a 4-fold increase in the liver expression of the LBP whereas prebiotic treatment decreased liver expression of LBP under HFD (Figure 8a). This is associated with the normalisation of the Myd88 expression in the liver (Figure 8b), a protein located downstream of LPS receptor, toll-like receptor 4, thereby suggesting a decrease in the LPS-induced inflammation in the liver. The liver was previously claimed as the main source of the variation in the concentration of circulating LBP, but a recent study also suggested a role for the adipose tissue in the production of LBP in obesity (Moreno-Navarrete et al., 2013). Accordingly, in our study, HFD-induced obesity is associated with an increased LBP expression also in the adipose tissue and prebiotics reduced HFD-induced LBP expression in the adipose tissue (Figure 8c). Moreover, we confirmed the reduction in inflammation following prebiotic treatment in other organs since the administration of prebiotics in HFD-fed mice normalised the expression of the inflammatory markers MCP1 and CD11c in the adipose tissue (Figures 8d and e). These two proteins correspond, respectively, to a cytokine involved in immune cell recruitment and a marker of the primary population of macrophages increased in obesity (Osborn and Olefsky, 2012).
Figure 8.
Prebiotic treatment decreased inflammation, fat mass development and insulin resistance associated with HFD-induced obesity. Inflammatory markers mRNA expression: (a) LPS binding protein (encoded by LBP) and (b) Myeloid differentiation primary response gene (88) (encoded by Myd88) in the liver; (c) LPS binding protein (encoded by LBP); (d) Monocyte chemoattractant protein 1 (encoded by MCP1) and (e) Cluster of differentiation 11c (encoded by CD11c) in the epididymal adipose tissue (EAT); (f) Plasma leptin (ng μl−1) in cava vein and (g) Insulin resistance index determined by multiplying the area under the curve (from 0 to 15 min) of blood glucose and plasma insulin following an oral glucose load (2 g glucose per kg of body weight) measured in control diet-fed mice (CT) (n=9), CT diet-fed mice treated with prebiotics (CT-Pre) (n=10), HFD-fed mice (HFD) (n=10) and HFD-fed mice treated with prebiotics (HFD-Pre) (n=10). Data are means±s.e.m. Data with different superscript letters are significantly different (P<0.05) according to a post hoc ANOVA one-way statistical analysis.
In accordance with our previous data, showing that prebiotic treatment reduces fat mass and body weight (Cani et al., 2007b; Everard et al., 2011, 2013), we also found in the present study that the administration of prebiotics reduced levels of plasma leptin. This hormone, produced by the adipose tissue and reflecting the adiposity, was four times lower in HFD-Pre compared with HFD, whereas HFD dramatically increased plasma leptin (20-fold increase) as compared with CT mice (Figure 8f). Notably, the insulin resistance index was also reduced after prebiotic treatment (Figure 8g), demonstrating the beneficial effects of gut microbiota modulations on glucose homeostasis in obesity.
Discussion
This study demonstrates that an HFD and administration of prebiotics profoundly modify host antimicrobial peptide production and is associated with changes in gut microbial composition. The HFD-induced obesity was associated with significant changes in gut microbiota, with an increased proportion of Firmicutes and a decreased amount of the phyla Bacteroidetes, Cyanobacteria and Verrucomicrobia (all Gram negative) as well as Tenericutes (which lack a cell wall). We hypothesise that, in response to HFD, the host may contribute to the shift in the Firmicutes/Bacteroidetes ratio by modulating the production of three specific antimicrobial peptides (Reg3g, Pla2g2 and Lyz1).
Interestingly, prebiotic treatment under HFD massively increased Reg3g mRNA expression (>50-fold), whereas the mRNA expressions of the two other peptides were not affected by prebiotics.
The presence of some bacteria may affect the expression of Reg3g, which in turn contributes to shape the bacterial community. Diet has an important role in this interaction, influencing both host and microbial metabolism. Among the bacteria significantly affected by prebiotic treatment, we identified A. muciniphila, which was previously shown to increase Reg3g expression (Everard et al., 2013). We cannot rule out that other specific taxa also contribute to the regulation of these antimicrobial peptides and, potentially, host metabolism. Importantly, here we identified several bacterial taxa whose abundance was inversely affected by HFD and prebiotic treatment. We postulate that these specific changes may contribute to the beneficial effects of prebiotics on host metabolism. For example, we observed that the proportion of Bilophila was significantly increased (7-fold) during HFD feeding, whereas prebiotic treatment reversed this trend (Supplementary Table S2). In accordance with this observation, it was recently demonstrated that Bilophila wadsworthia emerges under pathological conditions such as appendicitis and other intestinal inflammatory disorders (Devkota et al., 2012). Bile acids are involved in the selection of this type of bacteria, and Bilophila wadsworthia metabolites might serve as mucosal barrier breakers that permit immune cell infiltration. Consistent with this hypothesis, we previously demonstrated that obesity and HFD are associated with gut barrier disruptions by a mechanism involving a decrease in mucus layer thickness (Everard et al., 2013), alterations in antimicrobial peptide production (Everard et al., 2013) and tight-junction protein delocalisation (Cani et al., 2009; Everard et al., 2012), whereas prebiotic treatment restores Reg3g expression (Figure 1) and improves the localisation and distribution of tight junctions (Cani et al., 2009). Histone deacetylases inhibition has been shown to increase Reg3g expression (Turgeon et al., 2013). Therefore, we may not rule out that the higher Reg3g expression observed upon prebiotic treatment may be related to the increased intestinal butyrate content, a well-known histone deacetylase inhibitor.
Among the bacteria potentially beneficial for host physiology, we identified the genus Allobaculum. This finding is consistent with a previous study showing that low-fat feeding was associated with an increase in the genus Allobaculum compared with HFD feeding (Ravussin et al., 2011). Interestingly, treatment with the plant alkaloid berberine, which prevents obesity and insulin resistance in rats fed an HFD, increased the abundance of Allobaculum (Zhang et al., 2012). Moreover, changes in the proportion of Bifidobacterium related to diet were observed in this study (Supplementary Table S2). We previously observed that HFD feeding decreases Bifidobacterium spp. (Cani et al., 2007a, 2007b) and that the abundance of these bacteria is inversely correlated with gut permeability, metabolic endotoxemia and low-grade inflammation, whereas prebiotic treatment improved these parameters (Cani et al., 2007a, 2007b). In line with a previous report (Ravussin et al., 2011), we found that HFD increased the proportion of Bacteroides and Mucispirillum. Our results show that Bacteroides were even more increased by prebiotic treatment, whereas Mucispirillum were not affected by the prebiotic treatment, thereby suggesting that these genera were not directly involved in the phenotype of these mice.
The observed effects of the HFD on the functional profile of the mouse caecal microbiota, that is, the increase in PTS, ABC transporter and cell motility functions, are in accordance with several previous studies. For example, PTS enzymes were enriched in human faecal microbiomes of obese and inflammatory bowel diseases patients (Greenblum et al., 2012). Similarly, Turnbaugh et al. (2009) demonstrated that feeding an high-fat carbohydrate diet in humanised gnotobiotic mice correlated with an enhanced proportion of PTS and ABC transporters. Furthermore, ‘Cell Motility' functions are enriched in the gut microbiomes of obese individuals (Ferrer et al., 2013). Among the pathways involved in cell motility, we observed that COGs involved in flagellar assembly pathway were generally increased upon HFD, whereas prebiotic treatment decreased their abundance in most cases. A similar association between the enrichment of the flagellar assembly pathway and an HFD was recently reported in obese adolescents (Ferrer et al., 2013). Here, we demonstrated that prebiotic supplementation under HFD decreased the proportion of many COGs related to PTS, ABC transporters and cell motility (Supplementary Table S3). Based on these observations, it is tempting to speculate that reducing the amount of bacteria harbouring putative agonists of toll-like receptor 5 signalling (that is, flagellin) contributed to decreased obesity observed in the prebiotic-treated group (Everard et al., 2013). However, the absence of toll-like receptor 5 is associated with metabolic syndrome development (Vijay-Kumar et al., 2010), suggesting that this connection requires further investigation.
In contrast to recent studies (Daniel et al., 2013; Le Chatelier et al., 2013), we observed that the proportion of COGs potentially associated with butyrate production was significantly increased during obesity. However, our observation is in accordance with previous studies suggesting that butyrate production is associated with body weight gain (Turnbaugh et al., 2006; Ferrer et al., 2013). Therefore, we measured the caecal SCFA content and we did not find any modification in caecal butyrate content under HFD compared with CT diet whereas prebiotics treatment clearly increased caecal butyrate content. This discrepancy between COGs analysis and SCFA measurement may be due to the fact that by the gut microbiome analysis we are not able to assess the complete pathway of SCFA synthesis and all the interaction occurring within it. In our study, the assessment of the metabolic potential relied on the counts of metagenomic fragments assigned to COGs. However, the amount of butyrate (and other metabolites) produced depends on the expression level (and activity) of relevant enzymes, which could be, in turn, altered by the diet type.
Importantly, taxa and functional gut microbiota modulations induced by prebiotics are associated with beneficial effects on obesity and related metabolic disorders such as a decrease in inflammation, a decrease in plasma leptin levels and an improvement of glucose homeostasis. The decreased plasma leptin observed in our study is in accordance with a recent elegant study investigation of the prebiotic treatment on the metabolomics profile in humanised gnotobiotic mice (Respondek et al., 2013).
Interestingly, the decrease in LBP production may be associated with a reduction in the sensitivity of cells to LPS and could represent a new mechanism involved in the reduction of inflammation induced by prebiotic in obesity.
Our data suggest that, under HFD, the host contributes to the modification of the microbial community by modulating the production of three specific antimicrobial peptides (Reg3g, Pla2g2 and Lyz1), which predominantly alter the viability of Gram-positive bacteria (Gallo and Hooper, 2012). Moreover, we observed that prebiotic treatment normalised the Firmicutes/Bacteroidetes ratio and increased the proportion of phyla Bacteroidetes, Tenericutes, Cyanobacteria and Verrucomicrobia phyla. We postulate that the Reg3g upregulation observed following prebiotic treatment may contribute to this effect because the expression of the other two peptides was not affected by prebiotic treatment. Consistent with this hypothesis, we observed that the abundance of several genes involved in LPS synthesis, which consequently represent Gram-negative markers, was decreased upon HFD (Supplementary Table S3). This observation is in agreement with specific changes in specific Gram-negative bacteria, as observed at the different taxonomic levels (that is, increased Bacteroidetes, Bacteroides and Akkermansia).
This study clearly demonstrated a lack of a direct relationship between the gut microbiota LPS synthesis potential and metabolic endotoxemia. A decrease in gut microbiome LPS biosynthetic pathway (in response to HFD) did not lead to lower LBP production, whereas an increase in gut microbiome LPS biosynthetic pathway does not lead to higher LBP production. One explanation for this counterintuitive result is that HFD increases gut permeability whereas prebiotic treatment restores gut barrier functions, increases intestinal epithelial cell turnover (that is, intectin) and decreases gut permeability.
Finally, our results demonstrate that the dietary interventions in mice may change gut microbiota richness and diversity at both the functional and taxonomic levels. A recent study in humans emphasised the importance of developing interventional procedures aimed at increasing gut microbiota richness for conditions such as obesity and metabolic disorders (Cotillard et al., 2013). In conclusion, through the use of metagenomic and physiological analyses, we discovered that prebiotic treatment profoundly affects numerous metabolic functions of the gut microbiota in obese and type 2 diabetic mice. In addition to being able to describe the functional capacities of the gut microbiota following prebiotic treatment upon control or diet-induced obesity and type 2 diabetes, we observed putative links between key taxa, metabolic processes of the gut microbiota and host antimicrobial peptide production. We uncovered unanticipated changes in metabolic processes during prebiotic treatment that may contribute to the improved phenotype observed in obese and type 2 diabetic mice treated with prebiotics. Thus, our results provide a foundation for the discovery of novel, interesting taxa or metabolic functions that are involved in the development of metabolic inflammation, gut barrier dysfunction and adipose tissue development associated with HFD feeding that can be counteracted by a prebiotic approach.
Acknowledgments
We wish to thank B Es Saadi, A Bever, A Barrois and R Selleslagh for technical assistance. PDC is a research associate at FRS-FNRS (Fonds de la Recherche Scientifique), Belgium. AE is a doctoral fellow at FRS-FNRS, Belgium. PDC is the recipient of FNRS and FRSM (Fonds de la recherche scientifique médicale, Belgium) and ARC (Action de Recherche Concertée). PDC is a recipient of ERC Starting Grant 2013 (European Research Council, Starting grant 336452-ENIGMO).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- Cani PD, Possemiers S, Van de WT, Guiot Y, Everard A, Rottier O, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe LJ, Humphreys NE, Lane TE, Potten CS, Booth C, Grencis RK. Accelerated intestinal epithelial cell turnover: a new mechanism of parasite expulsion. Science. 2005;308:1463–1465. doi: 10.1126/science.1108661. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- Daniel H, Gholami AM, Berry D, Desmarchelier C, Hahne H, Loh G, et al. High-fat diet alters gut microbiota physiology in mice. ISME J. 2013;8:295–308. doi: 10.1038/ismej.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewulf EM, Cani PD, Claus SP, Fuentes S, Puylaert PG, Neyrinck AM, et al. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut. 2013;62:1112–1121. doi: 10.1136/gutjnl-2012-303304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GM, Neyrinck AM, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes. 2011;60:2775–2786. doi: 10.2337/db11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Geurts L, Van Roye M, Delzenne NM, Cani PD. Tetrahydro iso-alpha acids from hops improve glucose homeostasis and reduce body weight gain and metabolic endotoxemia in high-fat diet-fed mice. PLoS One. 2012;7:e33858. doi: 10.1371/journal.pone.0033858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Cani PD. Diabetes, obesity and gut microbiota. Best Pract Res Clin Gastroenterol. 2013;27:73–83. doi: 10.1016/j.bpg.2013.03.007. [DOI] [PubMed] [Google Scholar]
- Fei N, Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013;7:880–884. doi: 10.1038/ismej.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M, Ruiz A, Lanza F, Haange SB, Oberbach A, Till H, et al. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Environ Microbiol. 2013;15:211–226. doi: 10.1111/j.1462-2920.2012.02845.x. [DOI] [PubMed] [Google Scholar]
- Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Quintela A, Alonso M, Campos J, Vizcaino L, Loidi L, Gude F. Determinants of serum concentrations of lipopolysaccharide-binding protein (LBP) in the adult population: the role of obesity. PLoS One. 2013;8:e54600. doi: 10.1371/journal.pone.0054600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci USA. 2012;109:594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner F. Studies with inulin-type fructans on intestinal infections, permeability, and inflammation. J Nutr. 2007;137:2568S–2571S. doi: 10.1093/jn/137.11.2568S. [DOI] [PubMed] [Google Scholar]
- Hailman E, Lichenstein HS, Wurfel MM, Miller DS, Johnson DA, Kelley M, et al. Lipopolysaccharide (LPS)-binding protein accelerates the binding of LPS to CD14. J Exp Med. 1994;179:269–277. doi: 10.1084/jem.179.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Kitazawa H, Nishihara T, Nambu T, Nishizawa H, Iwaki M, Fukuhara A, et al. Intectin, a novel small intestine-specific glycosylphosphatidylinositol-anchored protein, accelerates apoptosis of intestinal epithelial cells. J Biol Chem. 2004;279:42867–42874. doi: 10.1074/jbc.M408047200. [DOI] [PubMed] [Google Scholar]
- Klimke W, Agarwala R, Badretdin A, Chetvernin S, Ciufo S, Fedorov B, et al. The National Center for Biotechnology Information's Protein Clusters Database. Nucl Acids Res. 2009;37:D216–D223. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Navarrete JM, Ortega F, Serino M, Luche E, Waget A, Pardo G, et al. Circulating lipopolysaccharide-binding protein (LBP) as a marker of obesity-related insulin resistance. Int J Obes (Lond) 2012;36:1442–1449. doi: 10.1038/ijo.2011.256. [DOI] [PubMed] [Google Scholar]
- Moreno-Navarrete JM, Escote X, Ortega F, Serino M, Campbell M, Michalski MC, et al. A role for adipocyte-derived lipopolysaccharide-binding protein in inflammation- and obesity-associated adipose tissue dysfunction. Diabetologia. 2013;56:2524–2537. doi: 10.1007/s00125-013-3015-9. [DOI] [PubMed] [Google Scholar]
- Muccioli GG, Naslain D, Backhed F, Reigstad CS, Lambert DM, Delzenne NM, et al. The endocannabinoid system links gut microbiota to adipogenesis. Mol Syst Biol. 2010;6:392. doi: 10.1038/msb.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med. 2012;18:363–374. doi: 10.1038/nm.2627. [DOI] [PubMed] [Google Scholar]
- Pott J, Hornef M. Innate immune signalling at the intestinal epithelium in homeostasis and disease. EMBO Rep. 2012;13:684–698. doi: 10.1038/embor.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, et al. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity (Silver Spring) 2011;20:738–747. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Respondek F, Gerard P, Bossis M, Boschat L, Bruneau A, Rabot S, et al. Short-chain fructo-oligosaccharides modulate intestinal microbiota and metabolic parameters of humanized gnotobiotic diet induced obesity mice. PLoS One. 2013;8:e71026. doi: 10.1371/journal.pone.0071026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho M, Tang H, Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucl Acids Res. 2010;38:e191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz AG, Casafont F, Crespo J, Cayon A, Mayorga M, Estebanez A, et al. Lipopolysaccharide-binding protein plasma levels and liver TNF-alpha gene expression in obese patients: evidence for the potential role of endotoxin in the pathogenesis of non-alcoholic steatohepatitis. Obes Surg. 2007;17:1374–1380. doi: 10.1007/s11695-007-9243-7. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS One. 2011;6:e17288. doi: 10.1371/journal.pone.0017288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Yu Z, Ye X, Zou S, Li H, Yu D, et al. A marker of endotoxemia is associated with obesity and related metabolic disorders in apparently healthy Chinese. Diabetes Care. 2010;33:1925–1932. doi: 10.2337/dc10-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Chen J, Li W, Altintas I, Lin A, Peltier S, et al. Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: the CAMERA resource. Nucl Acids Res. 2011;39:D546–D551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Turgeon N, Blais M, Gagne JM, Tardif V, Boudreau F, Perreault N, et al. HDAC1 and HDAC2 restrain the intestinal inflammatory response by regulating intestinal epithelial cell differentiation. PLoS One. 2013;8:e73785. doi: 10.1371/journal.pone.0073785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereecke L, Beyaert R, van Loo G. Enterocyte death and intestinal barrier maintenance in homeostasis and disease. Trends Mol Med. 2011;17:584–593. doi: 10.1016/j.molmed.2011.05.011. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann A, Allahyar A, Greiner TU, Plovier H, Lunden GO, Larsson T, et al. Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host Microbe. 2013;14:582–590. doi: 10.1016/j.chom.2013.09.012. [DOI] [PubMed] [Google Scholar]
- Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Fei N, Pang X, Shen J, Wang L, Zhang B, et al. 2013A gut microbiota-targeted dietary intervention for amelioration of chronic inflammation underlying metabolic syndrome FEMS Microbiol Ecol; e-pub ahead of print 30 september 2013; doi: 10.1111/1574-6941.12228 [DOI] [PMC free article] [PubMed]
- Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, et al. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS One. 2012;7:e42529. doi: 10.1371/journal.pone.0042529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.