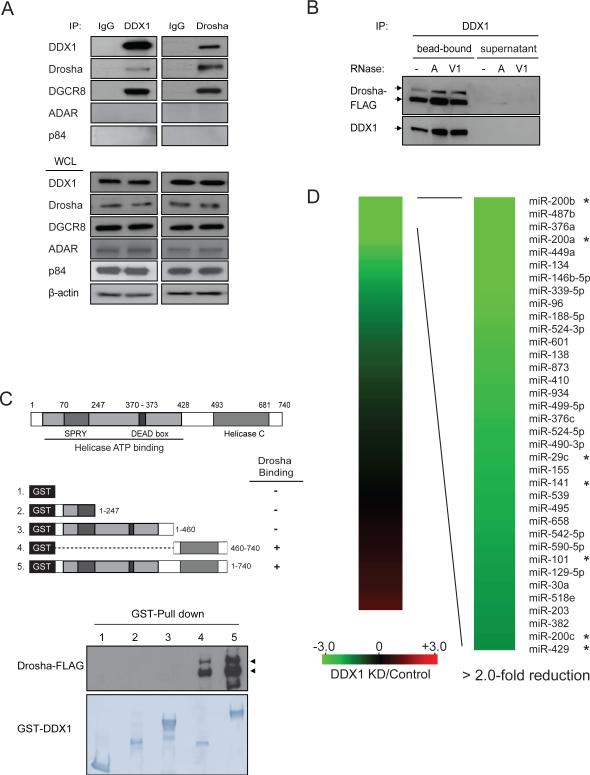
Figure 1. Interaction of DDX1 with Drosha microprocessor.
(A) Interaction between endogenous DDX1 and Drosha/DGCR8. Immunoprecipitation (IP) and Western blotting analyses were performed using indicated antibodies. Normal IgG was used as a negative control for IP. RNA-binding proteins ADAR and p84 were used as negative controls for the Drosha-binding proteins. WCL: whole cell lysate. (B) RNA-independent interaction between DDX1 and Drosha. DDX1 immunoprecipitates were treated with RNase A (10U) or RNase V1 (10U) and separated to supernatant and bead-bound fractions for Western blotting analyses. (C) Schematic representation of DDX1 domains (upper panel) and the interaction between Drosha and truncated forms of DDX1 (bottom panel). GST-fused DDX1 proteins were immobilized on glutathione Sepharose beads and mixed with the lysate of HEK293T cells expressing Drosha-FLAG. (D) Depletion of DDX1 inhibits the expression of a subset of miRNAs. Total RNAs from control and DDX1-knockdown (KD) were subject to global miRNA profiling analyses using q-PCR miRNA microarray. Green or red color on the heat map indicates a decrease or increase of miRNA level, and color intensities correspond to relative signal levels. Thirty-six miRNAs with over 2-fold reduction in the DDX1-knockdown cells were identified as DDX1-dependent miRNAs. The DDX1-dependent miRNAs in the miR-200 family and the 8-miRNA signature for mesenchymal subtype of ovarian cancer were marked with *. See also Figure S1.