Abstract
The routine use of genomic sequencing in clinical medicine has the potential to dramatically alter patient care and medical outcomes. To fully understand the psychosocial and behavioral impact of sequencing integration into clinical practice, it is imperative that we identify the factors that influence sequencing-related decision making and patient outcomes. In an effort to develop a collaborative and conceptually grounded approach to studying sequencing adoption, members of the National Human Genome Research Institute's Clinical Sequencing Exploratory Research Consortium formed the Outcomes and Measures Working Group. Here we highlight the priority areas of investigation and psychosocial and behavioral outcomes identified by the Working Group. We also review some of the anticipated challenges to measurement in social and behavioral research related to genomic sequencing; opportunities for instrument development; and the importance of qualitative, quantitative, and mixed-method approaches. This work represents the early, shared efforts of multiple research teams as we strive to understand individuals' experiences with genomic sequencing. The resulting body of knowledge will guide recommendations for the optimal use of sequencing in clinical practice.
Keywords: behavior, genome sequencing, measures, outcomes, psychosocial
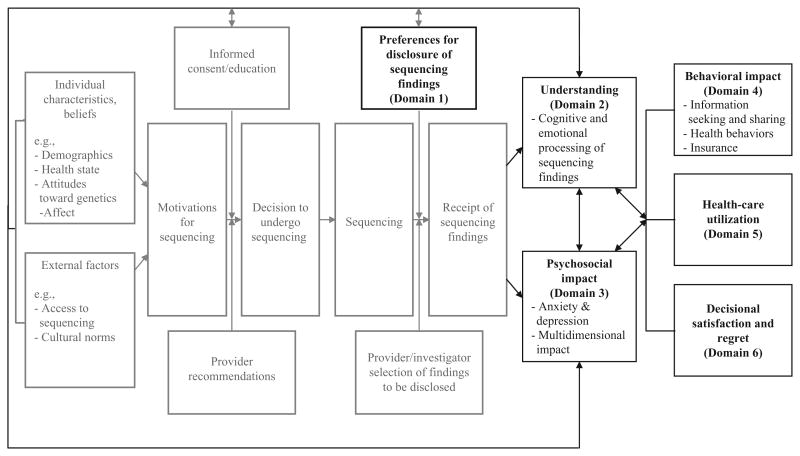
The dramatic reduction in the cost of genomic sequencing coupled with the improved accuracy of genomic technologies has set the stage for routine use of whole-exome and whole-genome sequencing in medical care. Although sequencing holds the potential to improve patient outcomes, models for the optimal delivery of genomic care are lacking. To systematically investigate the impact of sequencing integration on individuals and health systems, and to foster collaboration in research on the major ethical, legal, and social implications of sequencing technologies, the National Human Genome Research Institute (NHGRI) formed the Clinical Sequencing Exploratory Research (CSER) Consortium. A principal challenge faced by CSER investigators is the need to accurately measure the factors that influence sequencing-related decision making and outcomes. To that end, investigators formed the Outcomes and Measures Working Group to harmonize some of the patient-centered outcome measures that will be used in CSER projects, provide a forum for discussing development of novel measures, and facilitate cross-study, data-driven analyses. Trough an iterative process, we have identified and shared knowledge about measures that are in the public domain and discussed the pros and cons of outcome measurement in a variety of areas. In some cases, we have reached consensus on the measures that might be appropriate for use across our diverse study settings and populations. Although our group has identified many high-priority domains for investigation (Figure 1), we do not endorse specific measures. In this article, we outline priority areas for ethical, legal, and social implications research that working group members have identified to date. Although some CSER sites are also investigating provider perspectives on sequencing, that work will not be the focus of this article. Rather, this research agenda serves as a first step in the development of a collaborative and conceptually grounded approach to studying participant outcomes in anticipation of future widespread sequencing adoption.
Figure 1. The process and potential outcomes of genomic sequencing.
Dark boxes are outcomes that are featured in this article. Gray boxes represent steps in the clinical process.
Overview of the CSER Consortium
The projects in the CSER Consortium investigate a diverse set of research questions in a variety of clinical and research contexts (Supplementary Table S1 online). Briefly, the CSER Consortium includes projects funded through multiple mechanisms by the NHGRI, including U01 projects (RFA-HG-10–017), R01 projects (RFA-HG-11-003), R21 projects (RFA-HG-11-004), and investigator-initiated grants, as well as an NHGRI Intramural sequencing study. The U01 projects evaluate the integration of genomic sequencing in the clinical care of healthy individuals and adults with cardiomyopathy (Brigham and Women's MedSeq); in adults with metastatic lung and colon cancer (Dana-Farber Cancer Institute/Broad CanSeq); in adults who have clinical indications for testing for colorectal cancer/polyposis (University of Washington); in adults with cancer and cardiac disorders, children with dysmorphic findings, and adults and children with neurological disorders (University of North Carolina); in pediatric cancer patients (Baylor College of Medicine); and in children with heterogeneous disorders (Children's Hospital of Philadelphia). The investigators of the R01, R21, and investigator-initiated projects are studying issues related to the moral (The Children's Mercy Hospital) and legal duty to return sequencing research results (in pediatrics at Vanderbilt University and in newborn screening at Johns Hopkins University); the standards for sequencing-related informed consent (Columbia University); research participants' preferences for the return of research sequencing results (Seattle Children's Hospital, Columbia University, and Boston Children's Hospital); strategies for offering incidental findings to biobank research participants and deceased research participants' family members (University of California, San Francisco/Mayo Clinic/University of Minnesota); and attitudes and beliefs of patients and genetics professionals regarding the return of diagnostic genomic findings (Case Western Reserve University). The NHGRI Intramural program funds sequencing research on well-phenotyped adults (ClinSeq). Although the NHGRI has recently funded additional clinical sequencing U01 sites that are now part of the Consortium, investigators from those projects were not involved in the early work of the Outcomes and Measures Working Group that is detailed in this article.
Domains of Interest and Measurement of Impact
The Outcomes and Measures Working Group identified six major domains for coordination (Table 1). We considered a number of key factors when evaluating the domains for coordination, including whether the domains (i) are included in health decision-making or health behavior models that are being used at individual CSER sites, (ii) are being evaluated by at least two CSER projects, (iii) are the subject of prior genomics work, and (iv) were identified through clinical observation. For each area, we have identified compelling reasons for inclusion of the domain in Consortium studies, a brief overview of the published literature, a general description of how Consortium projects are addressing the domain, and challenges to domain measurement. Importantly, we recognize that measuring the impact of genomic sequencing is dependent on study design. Experimental designs—i.e., randomized controlled trials comparing patients who receive genomic sequencing with those who do not—may disentangle the effect of potential confounding factors. Observational studies (i.e., non–randomized controlled trials) may provide more descriptive data about the impact of genomic sequencing on individuals. Both study designs have advantages and disadvantages and are represented across the various projects in the Consortium.
Table 1. Outcome measures coordinated across CSER sites by domain.
| Selected outcomes included in CSER projectsa |
|---|
| Preferences |
| Preferred role of patient in medical decision making7 |
| Novel preference measures under development by CSER sites |
| Elicitation of sequencing-related preferences through hypothetical vignettes |
| Elicitation of actual preferences for the disclosure of incidental information |
| Elicitation of preferences for the integration of sequencing results in medical records |
|
|
| Understanding |
| Knowledge about genetics and genome sequencing16 |
| Novel understanding measures under development by CSER sites |
| Understanding of sequencing-related informed consent concepts |
| Comprehension of the implications of sequencing findings |
| Recall of sequencing results disclosed |
| Perceptions of uncertainty related to sequence information |
|
|
| Psychosocial impact |
| Anxiety and depression28–30,72 |
| Multidimensional impact34,35 |
|
|
| Behavioral impact |
| Information seeking & sharing73, 74 |
| Health information engagement and apprehension |
| Communication of sequencing findings to family members |
| Novel information seeking and sharing measures under development by CSER sites |
| Health behavior |
| Physical activity |
| Fruit and vegetable consumption |
| Smoking |
| Novel behavior measures under development by CSER sites (examples) |
| Changes in medical treatment |
| Vitamin, supplement, and medication use |
| Increased/decreased motivation to enact a variety of health-related behaviors |
|
|
| Health-care utilization |
| Hospital utilization and access to care52 |
| Novel health-care utilization measures under development by CSER sites (examples) |
| Medical care visits (e.g., general practitioner, medical specialist, genetics provider) |
| Hospital-based care (e.g., acute care, planned care) |
| Medical tests/procedures (e.g., screening, diagnostic, surveillance) |
| Medication changes (e.g., prescription, over the counter) |
| Insurance (e.g., health, life, disability, long-term care) |
| Health behavior programs (e.g., smoking cessation) |
|
|
| Decisional satisfaction & regret |
| Decision regret66 |
| Decisional conflict75 |
| Novel decision satisfaction and regret measures under development by CSER sites |
| Satisfaction with communication of sequencing results |
| Satisfaction with sequencing results |
| Sequencing-related expectations satisfied |
CSER, Clinical Sequencing Exploratory Research Consortium.
Only measures that are utilized by two or more sites in the consortium are referenced. Citations do not provide a complete list of measures available for these domains, nor are they intended to be an endorsement of particular measures as the optimal measures in that domain.
Conceptual Domains
Preferences for information
There is much debate about what role patient preferences should play in the disclosure of genomic sequencing results. When asked hypothetically, most patients and research participants express interest in receiving all types of results, even those of uncertain signifcance.1 Facio et al.1 reported that 95% of participants in the NHGRI ClinSeq study wished to learn results from whole-genome sequencing and that intentions to learn results were higher for actionable findings and carrier status as compared with nonactionable findings and uncertain genomic results.1 However, despite this general enthusiasm, the uptake of clinical genetic testing in the setting of single-gene studies in some cases is considerably lower,2,3 suggesting that a subset of at-risk individuals hesitate to receive genetic information. Evidence from decision-making research demonstrates that intentions predict only a subset of health-related behaviors.4 In addition, genetic testing uptake may be influenced by factors such as access to testing, insurance coverage, and provider knowledge.2
In July 2013, the American College of Medical Genetics and Genomics issued a recommendation that all laboratories conducting genomic sequencing seek out pathogenic or likely pathogenic variants in 56 genes associated with actionable conditions and that the results be disclosed to the ordering physician, irrespective of patient preferences.5 These recommendations have generated substantial controversy. Although some Consortium projects do not rely on individual preferences to inform decisions about the return of results, others are specifically designed to assess and accommodate individual preferences.
A principal challenge in measuring preferences, whether hypothetically or in the context of actual decision making, is that there are few validated measures of preferences for the disclosure of genomic findings. Broader measures of preferences for shared decision making, such as the Shared Decision Making Questionnaire6 and the Control Preference Scale,7 provide insight into what roles participants would like to play in decisions about the communication of genetic test information. However, many projects are developing novel preference measures, posing questions about individuals' desires for information categorized according to key attributes (e.g., actionable versus nonactionable) and/or medical indications (e.g., pharmacogenomics, disease risk alleles). Qualitative methods can provide a nuanced understanding of individual preferences, by allowing investigators to probe participants on how they understand constructs such as actionability and uncertainty. Finally, many Consortium studies have decided that it is important to assess preferences for results both pre- and postdisclosure, in order to assess the stability of individual preferences over time and to determine the impact of disclosure on preference stability.
Participant understanding: cognitive and emotional processing of sequencing findings
The Working Group devoted considerable attention to the challenges of measuring how individuals understand and process information in the context of sequencing. “Understanding” in the setting of genomic sequencing can be conceptualized in many ways and assessed at multiple points in the testing process. Furthermore, there is a lack of consensus across CSER sites about how understanding should be defined and operationalized. Although assessment of baseline understanding (e.g., genetic knowledge), understanding after an informed consent session, and understanding of the limitations of sequencing technologies are essential—and are therefore key independent variables in a number of projects—we focus the present discussion on the understanding of disclosed sequencing results (e.g., health implications of testing). Accurately measuring how individuals process and understand disclosed sequencing information is imperative because it allows us to evaluate the adequacy of existing systems for the return of genomic findings.
Prior studies have assessed individuals' knowledge, risk perception, and information recall in the setting of genetic testing. Disclosure of genetic test results can lead to more accurate risk perception and increased knowledge.8 Moreover, a study of multiplex genetic susceptibility testing concluded that patients commonly recall test results correctly and do not interpret test results in overly deterministic ways.9 Yet disclosure may also be associated with misinterpretation and confusion. For example, individuals who receive “negative” results can underestimate their risk to pass a condition onto a child,10 and individuals who are informed that they have variants of unknown significance or intermediate-risk alleles may have difficulty understanding and interpreting their results.10–12 Individuals can also maintain pretesting risk perceptions, even when they understand the implications of genomic test results. For example, a study of APOE testing for Alzheimer disease risk showed that a subset of research participants accurately recalled their testing-based risk but still believed that their risk was either higher or lower than the risk provided by the study team.13
Measuring how individuals understand and process disclosed genomic sequencing information is challenging for several reasons. First, sequencing results vary in terms of actionability, predictive value, and potential impact.14 Measurement of understanding and information processing may be complicated by the fact that individuals often receive multiple results simultaneously and that the implications of each result may differ. In addition, there are different ways to assess accurate understanding. For example, should the gold standard for understanding be concordance between the person who disclosed the findings and the person whose DNA was sequenced? Should we measure the degree of understanding by asking individuals to report the health implications of testing and compare their answers to relevant data in expertly curated genomic databases? Is it enough that individuals accurately recall the information that was given to them, or should they also appreciate the personal relevance of the information? What weight should be given to subjective understanding (i.e., how well the individual believes he/she understands the information)?15
Projects in the CSER Consortium are measuring understanding, information processing, and affective response in a variety of ways. There are a few validated knowledge measures that are being adapted by some CSER Consortium projects, including a knowledge of sequencing scale developed by Kaphingst16 in conjunction with ClinSeq colleagues.9 Some sites have adapted a previously published measure of subjective understanding developed by Joffe et al.15 Given the fact that emotion plays an extremely important role in information processing and decision making,17,18 a number of sites are also using measures that include an assessment of affective outcomes such as anticipated regret and tolerance of uncertainty. The ClinSeq group is developing a novel measure of uncertainty, specific to genomic sequencing, that also includes an assessment of affective outcomes. Given the lack of standardization and complexities in defining and measuring postdisclosure understanding, however, the majority of projects have developed novel scales and are using qualitative methods (e.g., discourse analysis of disclosure visits, postdisclosure patient interviews) to explore the different ways individuals process and make meaning of information. These methods may help lay the groundwork for future measure development.
Psychological responses to the return of results
Research consistently shows that individuals generally experience minimal adverse psychological sequelae after receipt of genetic risk information. Even among individuals receiving results indicating increased disease susceptibility, most studies have shown either no change or a decrease in negative emotions such as anxiety and depression as compared with prereturn levels. These patterns are seen both in testing for conditions with no preventive options, such as Huntington disease, and in testing for conditions with preventive options, such as melanoma.19,20 Similarly, most individuals have benign emotional responses to single-nucleotide polymorphism analyses, providing genetic risk information about multiple conditions simultaneously.21 A possible exception to this general principle may occur among individuals with elevated psychological distress at baseline (representing a potentially more vulnerable group) and among certain individuals affected by disease as compared with healthy individuals.22–24
Although findings from prior work in the setting of single-gene and single-nucleotide polymorphism testing are generally reassuring, the effects of returning genomic sequencing results, particularly in a clinical context, are unexplored. Responses to the return of sequence results may differ for several reasons. First, genomic sequencing can provide huge quantities of unexpected risk information about a vast array of diseases. It may also provide information that is qualitatively different from susceptibility information, such as findings related to pharmacogenomics or ancestry. In other contexts, the disclosure of unexpected information has produced anxiety in some populations.25
Second, uncertainty pervades genomic sequencing information. Sequencing has the potential to introduce uncertainty not only about the probability of illness through the disclosure of known risk variants but also about whether specific variants are actually associated with disease risk at all (variants of uncertain significance).26 Unlike previous sources of genetic risk information, sequencing further introduces the complexity of uncertainty in disentangling gene–gene interactions and gene–environment interactions that lead to disease risk. Helping patients to understand these sources of uncertainty will be critical to managing expectations about sequencing information. To this end, ClinSeq investigators have developed and validated a novel scale of perceptions of uncertainty related to genomic sequencing that includes three subscales: medical, affective, and trustworthiness. Convergent and divergent validity data using measures of resilience and ambiguity aversion support hypothesized relationships. Publication of the scale is underway. Prior research suggests that providers may interpret uncertainty differently than do patients.27 Thus, effective communication will entail assessing perceptions of uncertainty to maximize understanding and minimize the negative impacts of uncertain information. Supplementary Table S2 online details some of the specific questions related to psychological outcomes that the CSER Consortium is positioned to address.
Numerous well-validated scales that assess emotional states and traits, including depression and anxiety, have been used in research on genetic testing.17 The majority of Consortium studies are using scales such as the 9-item Patient Health Questionnaire28 to measure depression, the 7-item General Anxiety Disorder scale29 to measure anxiety, or the 14-item Hospital and Depression Scale30 to measure both. In addition, well-established scales to measure more encompassing constructs such as quality of life and happiness may be more appropriate for specific studies.
Identifying sensitive instruments that focus specifically on psychological responses to genetic information has been more challenging. Although a number of scales have been developed and tailored for BRCA testing,31 Lynch syndrome testing,32 and APOE genotyping for Alzheimer disease risk,33 the Consortium has focused on two that can be adapted more easily to the disclosure of sequencing results. The first is the Psychological Adaptation to Genetic Information Scale,34 a 26-item instrument that assesses frequency of intrusive thoughts, ability to discuss results with others, self-worth, certainty about information, and perceived control over consequences of genetic information. The second is the Multidimensional Impact of Cancer Risk Assessment scale,35 a 25-item instrument assessing distress, uncertainty, and positive responses to genetic test results for cancer risk. Although both scales enhance the ability to examine the impact of specific information and more nuanced outcomes, each has limitations. Disadvantages of the Psychological Adaptation to Genetic Information Scale are limited use to date and an initial focus on single-gene testing, whereas disadvantages of the Multidimensional Impact of Cancer Risk Assessment include limited use outside of testing for cancer risk, focus on single-gene testing, and questionable internal consistency on subscales in some studies.36,37 Examining the performance of well-adapted versions of each of these scales in the CSER Consortium will provide valuable guidance for future research.
Adaptation of these scales also raises important conceptual questions. First, what impact are researchers trying to assess? Is it the impact of information disclosure generally or of disclosure of specific results? Second, in the latter case, how should psychological impact be measured when multiple results, and/or results that have implications for both the individual and his/her family, are returned? The Psychological Adaptation to Genetic Information Scale and Multidimensional Impact of Cancer Risk Assessment were developed for testing specific genes and thus provide results that can be more easily categorized as “mutation positive,” “mutation negative,” and “indeterminate.” More complex results may require broader conceptualization of potential responses. Third, when is the appropriate time to measure psychological responses? Data from targeted testing performed on largely self-selected populations have generally shown the emotional impact of test results to be transient.38–40 In situations such as when sequencing is being used to inform cancer care, individuals may not be able to appreciate the implications of sequencing results until treatments have been completed. Conversely, the emotional impact of learning about increased risk for future illness may intensify as individuals approach the typical age of onset or show initial signs of disease.41 Investigators will need to address these issues as they consider how to best adapt and administer questionnaires for their specific research questions, populations, and settings.
Finally, questions remain about whether mild “negative” psychological responses might be beneficial in some situations. Negative emotions can be powerful motivators for action when avenues exist to reduce risks, and efforts to minimize anxiety and distress may be counterproductive in situations in which patients would optimally engage with the threatening information and work toward reducing their risks.42
Behavioral impact
The CSER Consortium projects are also investigating the influence of genomic sequencing on various health behaviors. For instance, some projects are studying whether learning about previously unknown health risks through sequencing will motivate (or demotivate)43 adults to reduce those risks through health-promoting lifestyle changes (e.g., diet, smoking cessation). The Consortium is also investigating effects on plans or intentions regarding future behaviors such as those involving childbearing or insurance purchases. In addition, there is a broad interest in information seeking (e.g., individuals' attempts to get additional information about sequencing and results) and sharing (e.g., their disclosure of results to other individuals). Understanding the effects of sequencing on behavioral outcomes is important because of their potential to guide clinical adoption of sequencing and to inform policies affecting public health and health services.
Most of the existing research has investigated these kinds of outcomes after targeted genetic testing. Many studies have focused on changes in smoking behaviors following return of results showing increased genetic risk for pulmonary or other diseases.44–46 Although some positive effects have been found for smoking cessation46 and quit attempts, null results are common. More broadly, a recent Cochrane review of research on behavioral effects of providing genetic risk information identified effects on self-reported diet and intentions to change behavior, but little or no effects on actual behavior change for smoking and physical activity.47,48 Notably, the included studies were generally of poor quality and underpowered to detect what are likely to be small effects. In addition, potential negative behavioral effects of providing genetic risk information have not been rigorously studied. It is therefore premature to draw conclusions about positive or negative effects of genetic risk information—especially information from genomic sequencing—on behavioral outcomes.
Motivated by the dearth of research and the expansive scope of genomic sequencing information, several Consortium projects are using qualitative and mixed methods to develop a rich understanding of participants' behavioral responses to sequencing. These approaches are well suited to discovering patterns of behaviors and their psychosocial correlates. In addition, Consortium projects are using quantitative research methods to study behavioral outcomes. Several institutions are using brief validated measures (e.g., the Rapid Assessment of Physical Activity)49 as well as questions from large national studies such as the National Cancer Institute's Health Information National Trends Survey50 or Cancer Care Outcomes Research and Surveillance Consortium51 or the Centers for Disease Control and Prevention's National Health and Nutrition Examination Survey52 and Behavioral Risk Factor Surveillance System.53 Because suitable measures did not exist for some outcomes of interest (e.g., medication/supplement use, information sharing), many Consortium projects have developed novel measures.
Future research would benefit from guidance from theoretical models of health decision making or health behavior change.54 For instance, theory will be useful in determining the circumstances under which sequencing may initiate a “teachable moment” capable of prompting behavior change.55 The Information–Motivation–Behavioral Skills model states that enacting certain complex behaviors requires a combination of learning information about them, motivation to enact them, and the behavioral skills necessary for doing so.56 Thus, although genomic results may be capable of motivating behavior change (e.g., as indicated by changes in intentions), that change may not occur if other crucial factors are missing (e.g., skills or resources required for behavior change). Research currently under way in the Consortium to assess barriers and facilitators of decision-making and behavior change after testing could provide additional information about the potential for sequencing to shape individuals' health behaviors.
Health-care resource utilization
A major question about the clinical implementation of genomic sequencing is its economic impact on patients and on the health-care system. Multiple issues arise when sequencing data are integrated into clinical care. First, the expected costs of data generation, interpretation, and reporting are substantial. Second, many institutions will need to develop or expand their infrastructures to deliver genomic-based care and to provide adequate follow-up for a range of incidental findings. Needed resources will be highly dependent on the clinical indication for testing and on policies and processes for return of incidental findings. Economic resources that should be considered include (i) institutional genomic review committees, if utilized; (ii) education and genetic counseling for patients; (iii) education for health-care providers; (iv) evaluation of patient preferences for incidental findings; and (v) the time required to return and discuss results with patients.
A more controversial issue is the downstream impact of genomic sequencing results on utilization of health-care resources. Some have argued that incidental findings, false-positive results, and results that are ambiguous could result in expensive and possibly unnecessary tests and consultations, and also lead to increased anxiety and other possible harms.57
Projects in the CSER Consortium are studying the economic impact of genomic sequencing in a variety of ways, primarily through measurement of health-care resource utilization (HRU). A number of the short- and intermediate-term HRU behaviors being measured in the CSER studies are shown in Table 1. These data can be collected using a variety of methods, including evaluation of electronic medical records, reimbursement claims, patient surveys, and patient diaries. Each method has strengths and weaknesses. If patients are all enrolled in the same health-care system with electronic medical records or claims data, evaluation will be more straightforward.58 One challenge—as with all economic evaluations—is balancing the resources and burden of collecting these data with the value they provide. Another consideration is how to evaluate the longer-term HRU impact of changes in health status resulting from interventions initiated by findings from genomic sequencing. Typically, decision analysis models are used to estimate such broad and longer-term clinical and economic effects.
Four challenges specific to assessing whether genomic sequencing is a justified use of health-care resource utilization are (i) assessing the HRU of family members who may elect to pursue genomic testing based on the index patient's findings; (ii) assessing the tremendous diversity of potential health impacts and associated HRU changes resulting from return of incidental findings; (iii) incorporating inherent value that patients place on receiving findings, independent of their clinical relevance (personal utility); and (iv) identifying appropriate comparator groups in order to access the effectiveness of changes in care. At a minimum, investigators should consider collecting information about whether patients have informed family members of their results. Further efforts to identify uptake of genomic testing or other medical care services initiated by sharing of results may be warranted but will be resource intensive. Given the variety of incidental findings that could be returned, it will not be possible to model all potential impacts of return of incidental findings. Approaches to assess the depth and breadth of this issue will need to be developed based on experience with ongoing research studies. Last, although the value that patients place on personal utility is not routinely considered in the development of clinical or reimbursement guidelines, personal utility is a patient-centered outcome that may drive much of genomic sequencing uptake and utilization.59 Approaches to measure personal utility, understand its importance, and consider it in policy development are needed. As a preliminary step in exploring personal utility, at least one project will conduct a discrete-choice experiment that can provide estimates of the value patients place on different types of incidental findings.60
Decision satisfaction and regret
Another area of inquiry within the CSER Consortium includes an exploration of how individuals arrive at, and reflect upon, their decisions related to the receipt of genome sequencing results. Elements of the decision making process include, but are not limited to, understanding, engagement, risk perception, worries, uncertainty perceptions, response efficacy, and attitudes toward learning results. Various scales have been used for several decades to assess both the process and outcomes of medical decision making,61 and several outcome scales have been used to assess the quality of decision making specifically in the context of genetic testing. To date, much of the research in genetic testing has focused on outcomes of decisions to undergo prenatal testing and testing for cancer risk.62–65
In most of the CSER Consortium projects, when decision making is studied, the focus is on decision outcomes, including informed choice, decision satisfaction and/or regret, and satisfaction with communication. Often, eligible individuals are asked to make decisions not only about having genomic testing performed but also about the types of incidental findings they wish to receive. Such decisions could result in learning useful, distressing, or unwanted information that has implications not only for the person tested but also for family members. Decision-quality assessments are key to understanding the overall value of sequencing results in both clinical and research contexts. A number of the projects in the CSER Consortium are using the Decision Regret Scale to evaluate postdecision outcomes.66 It includes five items to rate the level of remorse or distress over a treatment-related decision. Decision regret has been shown to be negatively associated with satisfaction with the decision-making process and positively associated with poorer health outcomes and decisional conflict.66
Projects in the Consortium are also measuring satisfaction with physician–patient communication and with genetic counseling.67 Although some projects have developed novel communication satisfaction measures, others are adapting items (e.g., from the Health Information National Trends Survey,50 the Roter Interaction Analysis System)68 or are using validated instruments such as the general communication subscale of the Quality of End-of-Life Communication measure.69 Due to the diversity of communication systems and genomic decision support in the CSER projects, some projects also aim to assess individuals' satisfaction with the usability of computer systems employed in the course of genomic sequencing.70,71 In addition, a number of sites are creating novel measures to evaluate individuals' satisfaction with testing and items that aim to capture whether genomic sequencing meets their expectations.
Assessments of decision quality are likely to be related to other process and outcome variables that are being measured by most of the CSER projects. For example, several projects are assessing the process and/or content of informed consent or return of results, as well as psychosocial outcomes of genomic sequencing, such as quality of life, anxiety, multidimensional impact, and coping. The quality of the decision is likely to be predictive of these types of outcome measures as well.
Discussion
In addition to identifying core domains for coordination, our Working Group has discussed a number of methodological and conceptual issues that are likely to transcend individual projects. As noted previously, because the landscape of genomic sequencing is underexplored, complementary qualitative and quantitative research methods may be needed to capture the breadth and depth of people's experiences with sequencing. Such mixed-methods approaches are likely to yield the most comprehensive understanding of impact. Second, there may be significant differences, both in terms of impact and in terms of the assessment of impact, when sequencing is performed in a clinical versus a research context. For example, obligations related to the return of incidental findings or the requirements for informed consent might differ depending on whether sequencing is performed for clinical or research purposes. Third, practical, normative, and ethical factors may influence decisions about whether or not to elicit individuals' preferences for genomic test result disclosure, and decisions about preference elicitation are likely to influence impact. Finally, when assessing the impact of genomic sequencing on individuals, it is important to note that there are likely to be interactions between individual, health-care system, and social context characteristics, and outcomes. Accurate measurement of important covariates will be essential if we are to understand the ways in which the impact of sequencing varies across different populations and settings.
Although the CSER Consortium Outcomes and Measures working group has identified a number of domains as high priority for investigation, we readily acknowledge that rapid advances in genome science create a dynamic environment in which the implications of testing will evolve. Behavioral and social scientists, legal and ethics scholars, clinical investigators, and clinicians will need to coevolve with genome scientists in order to ensure that their work continually addresses core genomic questions. Continued efforts to coordinate sequencing-related research activities across sites and disciplines will aid in our understanding of individuals' experiences with genomic sequencing and help to establish best practices for sequencing programs of the future.
Supplementary Material
Acknowledgments
This work was funded by the National Human Genome Research Institute grants 1U01HG006485, 1U01HG006487, 1U01HG006492, 1U01HG006500, 1U01HG006507, 1U01HG005211, 1U01HG006546, 1U01HG007307, U01HG 007303, 5R01HG006618, 5R01HG006615, 5R01HG006600, 5R21HG006596, 5R21HG006612, 5R21HG006613, 5R21HG 006594, 1F32HG006993, and R01 GM109743, Rettie AE; the Intramural Research Program of the National Human Research Institute, National Institutes of Health; and the American Cancer Society (120529-MRSG-11-006-01-CPPB).
Footnotes
A complete list of secondary authors from the CSER Outcomes and Measures Working Group is provided in the Supplementary Appendix online.
Supplementary Material: Supplementary material is linked to the online version of the paper at http://www.nature.com/gim
Disclosure: S.J. was a member of a Data Safety Monitoring Board for Gen-zyme/Sanofi until November 2012. The other authors declare no conflict of interest.
References
- 1.Facio FM, Eidem H, Fisher T, et al. Intentions to receive individual results from whole-genome sequencing among participants in the ClinSeq study. Eur J Hum Genet. 2013;21:261–265. doi: 10.1038/ejhg.2012.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lerman C, Shields AE. Genetic testing for cancer susceptibility: the promise and the pitfalls. Nat Rev Cancer. 2004;4:235–241. doi: 10.1038/nrc1301. [DOI] [PubMed] [Google Scholar]
- 3.Sanderson SC, O'Neill SC, Bastian LA, Bepler G, McBride CM. What can interest tell us about uptake of genetic testing? Intention and behavior amongst smokers related to patients with lung cancer. Public Health Genomics. 2010;13:116–124. doi: 10.1159/000226595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagger MS, Chatzisarantis NL. Integrating the theory of planned behaviour and self-determination theory in health behaviour: a meta-analysis. Br J Health Psychol. 2009;14(Pt 2):275–302. doi: 10.1348/135910708X373959. [DOI] [PubMed] [Google Scholar]
- 5.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon D, Schorr G, Wirtz M, et al. Development and first validation of the shared decision-making questionnaire (SDM-Q) Patient Educ Couns. 2006;63:319–327. doi: 10.1016/j.pec.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Degner LF, Sloan JA, Venkatesh P. The Control Preferences Scale. Can J Nurs Res. 1997;29:21–43. [PubMed] [Google Scholar]
- 8.Hilgart JS, Coles B, Iredale R. Cancer genetic risk assessment for individuals at risk of familial breast cancer. Cochrane Database Syst Rev. 2012;2:CD003721. doi: 10.1002/14651858.CD003721.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaphingst KA, McBride CM, Wade C, et al. Patients' understanding of and responses to multiplex genetic susceptibility test results. Genet Med. 2012;14:681–687. doi: 10.1038/gim.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bekker H, Denniss G, Modell M, Bobrow M, Marteau T. The impact of population based screening for carriers of cystic fibrosis. J Med Genet. 1994;31:364–368. doi: 10.1136/jmg.31.5.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reiff M, Bernhardt BA, Mulchandani S, et al. What does it mean?: uncertainties in understanding results of chromosomal microarray testing. Genet Med. 2012;14:250–258. doi: 10.1038/gim.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semaka A, Balneaves LG, Hayden MR. Grasping the grey: patient understanding and interpretation of an intermediate allele predictive test result for Huntington disease. J Genet Couns. 2013;22:200–217. doi: 10.1007/s10897-012-9533-7. [DOI] [PubMed] [Google Scholar]
- 13.Linnenbringer E, Roberts JS, Hiraki S, Cupples LA, Green RC. I know what you told me, but this is what I think: perceived risk of Alzheimer disease among individuals who accurately recall their genetics-based risk estimate. Genet Med. 2010;12:219–227. doi: 10.1097/GIM.0b013e3181cef9e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biesecker LG. Opportunities and challenges for the integration of massively parallel genomic sequencing into clinical practice: lessons from the ClinSeq project. Genet Med. 2012;14:393–398. doi: 10.1038/gim.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joffe S, Cook EF, Cleary PD, Clark JW, Weeks JC. Quality of informed consent: a new measure of understanding among research subjects. J Natl Cancer Inst. 2001;93:139–147. doi: 10.1093/jnci/93.2.139. [DOI] [PubMed] [Google Scholar]
- 16.Kaphingst KA, Facio FM, Cheng MR, et al. Effects of informed consent for individual genome sequencing on relevant knowledge. Clin Genet. 2012;82:408–415. doi: 10.1111/j.1399-0004.2012.01909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finucane ML, Alhakami A, Slovic P, Johnson SM. The affect heuristic in judgments of risks and benefits. J Behav Decis Mak. 2000;13(1):1–17. [Google Scholar]
- 18.Damasio A. Descartes' Error: Emotion, Reason, and the Human Brain. Vintage; London: 2006. [Google Scholar]
- 19.Broadstock M, Michie S, Marteau T. Psychological consequences of predictive genetic testing: a systematic review. Eur J Hum Genet. 2000;8:731–738. doi: 10.1038/sj.ejhg.5200532. [DOI] [PubMed] [Google Scholar]
- 20.Kasparian NA, Meiser B, Butow PN, Simpson JM, Mann GJ. Genetic testing for melanoma risk: a prospective cohort study of uptake and outcomes among Australian families. Genet Med. 2009;11:265–278. doi: 10.1097/GIM.0b013e3181993175. [DOI] [PubMed] [Google Scholar]
- 21.Bloss CS, Schork NJ, Topol EJ. Effect of direct-to-consumer genomewide profiling to assess disease risk. N Engl J Med. 2011;364:524–534. doi: 10.1056/NEJMoa1011893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duisterhof M, Trijsburg RW, Niermeijer MF, Roos RA, Tibben A. Psychological studies in Huntington's disease: making up the balance. J Med Genet. 2001;38:852–861. doi: 10.1136/jmg.38.12.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michie S, Bobrow M, Marteau TM. Predictive genetic testing in children and adults: a study of emotional impact. J Med Genet. 2001;38:519–526. doi: 10.1136/jmg.38.8.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorval M, Patenaude AF, Schneider KA, et al. Anticipated versus actual emotional reactions to disclosure of results of genetic tests for cancer susceptibility: findings from p53 and BRCA1 testing programs. J Clin Oncol. 2000;18:2135–2142. doi: 10.1200/JCO.2000.18.10.2135. [DOI] [PubMed] [Google Scholar]
- 25.Ahman A, Runestam K, Sarkadi A. Did I really want to know this? Pregnant women's reaction to detection of a soft marker during ultrasound screening. Patient Educ Couns. 2010;81:87–93. doi: 10.1016/j.pec.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 26.Han PK, Klein WM, Arora NK. Varieties of uncertainty in health care: a conceptual taxonomy. Med Decis Making. 2011;31:828–838. doi: 10.1177/0272989X11393976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han PK. Conceptual, methodological, and ethical problems in communicating uncertainty in clinical evidence. Med Care Res Rev. 2013;70(suppl 1):14S–36S. doi: 10.1177/1077558712459361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroenke K, Spitzer RL, Williams JB. The PHQ-9: validity of a brief depression severity measure. J Gen Intern Med. 2001;16:606–613. doi: 10.1046/j.1525-1497.2001.016009606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spitzer RL, Kroenke K, Williams JB, Löwe B. A brief measure for assessing generalized anxiety disorder: the GAD-7. Arch Intern Med. 2006;166:1092–1097. doi: 10.1001/archinte.166.10.1092. [DOI] [PubMed] [Google Scholar]
- 30.Snaith RP, Zigmond AS. The hospital anxiety and depression scale. Br Med J (Clin Res Ed) 1986;292:344. doi: 10.1136/bmj.292.6516.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esplen MJ, Stuckless N, Hunter J, et al. The BRCA Self-Concept Scale: a new instrument to measure self-concept in BRCA1/2 mutation carriers. Psychooncology. 2009;18:1216–1229. doi: 10.1002/pon.1498. [DOI] [PubMed] [Google Scholar]
- 32.Esplen MJ, Stuckless N, Gallinger S, et al. Development and validation of an instrument to measure the impact of genetic testing on self-concept in Lynch syndrome. Clin Genet. 2011;80:415–423. doi: 10.1111/j.1399-0004.2011.01770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung WW, Chen CA, Cupples LA, et al. A new scale measuring psychologic impact of genetic susceptibility testing for Alzheimer disease. Alzheimer Dis Assoc Disord. 2009;23:50–56. doi: 10.1097/wad.0b013e318188429e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Read CY, Perry DJ, Duffy ME. Design and psychometric evaluation of the Psychological Adaptation to Genetic Information Scale. J Nurs Scholarsh. 2005;37:203–208. doi: 10.1111/j.1547-5069.2005.00036.x. [DOI] [PubMed] [Google Scholar]
- 35.Cella D, Hughes C, Peterman A, et al. A brief assessment of concerns associated with genetic testing for cancer: the Multidimensional Impact of Cancer Risk Assessment (MICRA) questionnaire. Health Psychol. 2002;21:564–572. [PubMed] [Google Scholar]
- 36.Shiloh S, Dagan E, Friedman I, Blank N, Friedman E. A follow-up study on men tested for BRCA1/BRCA2 mutations: impacts and coping processes. Psychooncology. 2013;22:417–425. doi: 10.1002/pon.2106. [DOI] [PubMed] [Google Scholar]
- 37.Aspinwall LG, Taber JM, Leaf SL, Kohlmann W, Leachman SA. Genetic testing for hereditary melanoma and pancreatic cancer: a longitudinal study of psychological outcome. Psychooncology. 2013;22:276–289. doi: 10.1002/pon.2080. [DOI] [PubMed] [Google Scholar]
- 38.Douma KF, Aaronson NK, Vasen HF, Bleiker EM. Psychosocial issues in genetic testing for familial adenomatous polyposis: a review of the literature. Psychooncology. 2008;17:737–745. doi: 10.1002/pon.1357. [DOI] [PubMed] [Google Scholar]
- 39.Heshka JT, Palleschi C, Howley H, Wilson B, Wells PS. A systematic review of perceived risks, psychological and behavioral impacts of genetic testing. Genet Med. 2008;10:19–32. doi: 10.1097/GIM.0b013e31815f524f. [DOI] [PubMed] [Google Scholar]
- 40.Rew L, Kaur M, McMillan A, Mackert M, Bonevac D. Systematic review of psychosocial benefits and harms of genetic testing. Issues Ment Health Nurs. 2010;31:631–645. doi: 10.3109/01612840.2010.510618. [DOI] [PubMed] [Google Scholar]
- 41.Paulsen JS, Hoth KF, Nehl C, Stierman L. The Huntington Study G. Critical periods of suicide risk in Huntington's disease. Am J Psychiatry. 2005;162:725–731. doi: 10.1176/appi.ajp.162.4.725. [DOI] [PubMed] [Google Scholar]
- 42.Witte K, Allen M. A meta-analysis of fear appeals: implications for effective public health campaigns. Health Educ Behav. 2000;27:591–615. doi: 10.1177/109019810002700506. [DOI] [PubMed] [Google Scholar]
- 43.Bates BR, Templeton A, Achter PJ, Harris TM, Condit CM. What does “a gene for heart disease” mean? A focus group study of public understandings of genetic risk factors. Am J Med Genet A. 2003;119A:156–161. doi: 10.1002/ajmg.a.20113. [DOI] [PubMed] [Google Scholar]
- 44.Audrain J, Boyd NR, Roth J, Main D, Caporaso NF, Lerman C. Genetic susceptibility testing in smoking-cessation treatment: one-year outcomes of a randomized trial. Addict Behav. 1997;22:741–751. doi: 10.1016/s0306-4603(97)00060-9. [DOI] [PubMed] [Google Scholar]
- 45.Hollands GJ, Whitwell SC, Parker RA, et al. Effect of communicating DNA based risk assessments for Crohn's disease on smoking cessation: randomised controlled trial. BMJ. 2012;345:e4708. doi: 10.1136/bmj.e4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McBride CM, Bepler G, Lipkus IM, et al. Incorporating genetic susceptibility feedback into a smoking cessation program for African-American smokers with low income. Cancer Epidemiol Biomarkers Prev. 2002;11:521–528. [PubMed] [Google Scholar]
- 47.Marteau TM, French DP, Griffin SJ. Effects of communicating DNA-based disease risk estimates on risk-reducing behaviours. Cochrane Database Syst Rev. 2010;(10):CD007275. doi: 10.1002/14651858.CD007275.pub2. [DOI] [PubMed] [Google Scholar]
- 48.Bize R, Burnand B, Mueller Y, Rège-Walther M, Camain JY, Cornuz J. Biomedical risk assessment as an aid for smoking cessation. Cochrane Database Syst Rev. 2012;(12):CD004705. doi: 10.1002/14651858.CD004705.pub4. [DOI] [PubMed] [Google Scholar]
- 49.Topolski TD, LoGerfo J, Patrick DL, Williams B, Walwick J, Patrick MB. The Rapid Assessment of Physical Activity (RAPA) among older adults. Prev Chronic Dis. 2006;3:A118. [PMC free article] [PubMed] [Google Scholar]
- 50.Nelson DE, Kreps GL, Hesse BW, et al. The Health Information National Trends Survey (HINTS): development, design, and dissemination. J Health Commun. 2004;9:443–60. doi: 10.1080/10810730490504233. discussion 81. [DOI] [PubMed] [Google Scholar]
- 51.Malin JL, Ko C, Ayanian JZ, et al. Understanding cancer patients' experience and outcomes: development and pilot study of the Cancer Care Outcomes Research and Surveillance patient survey. Support Care Cancer. 2006;14:837–848. doi: 10.1007/s00520-005-0902-8. [DOI] [PubMed] [Google Scholar]
- 52.Centers for Disease Control and Prevention (CDC); National Center for Health Statistics (NCHS) Department of Health and Human Services, Centers for Disease Control and Prevention; Hyattsville, MD: 2013. [Accessed January 16th 2014]. National Health and Nutrition Examination Survey Questionnaire (or Examination Protocol, or Laboratory Protocol) http://www.cdc.gov/nchs/nhanes.htm. [Google Scholar]
- 53.Centers for Disease Control and Prevention (CDC) U.S. Department of Health and Human Services, Centers for Disease Control and Prevention; Atlanta, GA: 2013. [Accessed January 16th 2014]. Behavioral Risk Factor Surveillance System Survey Questionnaire. http://www.cdc.gov/brfss/data_documentation/index.htm. [Google Scholar]
- 54.Glanz K, Bishop DB. The role of behavioral science theory in development and implementation of public health interventions. Annu Rev Public Health. 2010;31:399–418. doi: 10.1146/annurev.publhealth.012809.103604. [DOI] [PubMed] [Google Scholar]
- 55.McBride CM, Emmons KM, Lipkus IM. Understanding the potential of teachable moments: the case of smoking cessation. Health Educ Res. 2003;18:156–170. doi: 10.1093/her/18.2.156. [DOI] [PubMed] [Google Scholar]
- 56.Fisher JD, Fisher WA. Changing AIDS-risk behavior. Psychol Bull. 1992;111:455–474. doi: 10.1037/0033-2909.111.3.455. [DOI] [PubMed] [Google Scholar]
- 57.McGuire AL, Burke W. An unwelcome side effect of direct-to-consumer personal genome testing: raiding the medical commons. JAMA. 2008;300:2669–2671. doi: 10.1001/jama.2008.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O'Sullivan AK, Thompson D, Drummond MF. Collection of health-economic data alongside clinical trials: is there a future for piggyback evaluations? Value Health. 2005;8:67–79. doi: 10.1111/j.1524-4733.2005.03065.x. [DOI] [PubMed] [Google Scholar]
- 59.Grosse SD, Wordsworth S, Payne K. Economic methods for valuing the outcomes of genetic testing: beyond cost-effectiveness analysis. Genet Med. 2008;10:648–654. doi: 10.1097/gim.0b013e3181837217. [DOI] [PubMed] [Google Scholar]
- 60.Regier DA, Friedman JM, Makela N, Ryan M, Marra CA. Valuing the benefit of diagnostic testing for genetic causes of idiopathic developmental disability: willingness to pay from families of affected children. Clin Genet. 2009;75:514–521. doi: 10.1111/j.1399-0004.2009.01193.x. [DOI] [PubMed] [Google Scholar]
- 61.Scholl I, Koelewijn-van Loon M, Sepucha K, et al. Measurement of shared decision making - a review of instruments. Z Evid Fortbild Qual Gesundhwes. 2011;105:313–324. doi: 10.1016/j.zefq.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 62.Dormandy E, Hooper R, Michie S, Marteau TM. Informed choice to undergo prenatal screening: a comparison of two hospitals conducting testing either as part of a routine visit or requiring a separate visit. J Med Screen. 2002;9:109–114. doi: 10.1136/jms.9.3.109. [DOI] [PubMed] [Google Scholar]
- 63.Michie S, Dormandy E, Marteau TM. Informed choice: understanding knowledge in the context of screening uptake. Patient Educ Couns. 2003;50:247–253. doi: 10.1016/s0738-3991(03)00044-2. [DOI] [PubMed] [Google Scholar]
- 64.Stacey D, Bennett CL, Barry MJ, et al. Decision aids for people facing health treatment or screening decisions. Cochrane Database Syst Rev. 2011;(10):CD001431. doi: 10.1002/14651858.CD001431.pub3. [DOI] [PubMed] [Google Scholar]
- 65.Kasparian NA, Wakefield CE, Meiser B. Assessment of psychosocial outcomes in genetic counseling research: an overview of available measurement scales. J Genet Couns. 2007;16:693–712. doi: 10.1007/s10897-007-9111-6. [DOI] [PubMed] [Google Scholar]
- 66.Brehaut JC, O'Connor AM, Wood TJ, et al. Validation of a decision regret scale. Med Decis Making. 2003;23:281–292. doi: 10.1177/0272989X03256005. [DOI] [PubMed] [Google Scholar]
- 67.Charles S, Kessler L, Stopfer JE, Domchek S, Halbert CH. Satisfaction with genetic counseling for BRCA1 and BRCA2 mutations among African American women. Patient Educ Couns. 2006;63:196–204. doi: 10.1016/j.pec.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 68.Roter D, Larson S. The Roter interaction analysis system (RIAS): utility and flexibility for analysis of medical interactions. Patient Educ Couns. 2002;46:243–251. doi: 10.1016/s0738-3991(02)00012-5. [DOI] [PubMed] [Google Scholar]
- 69.Engelberg R, Downey L, Curtis JR. Psychometric characteristics of a quality of communication questionnaire assessing communication about end-of-life care. J Palliat Med. 2006;9:1086–1098. doi: 10.1089/jpm.2006.9.1086. [DOI] [PubMed] [Google Scholar]
- 70.Lewis JR. IBM computer usability satisfaction questionnaires: psychometric evaluation and instructions for use. Int J Hum Comput Interact. 1995;7(1):57–78. [Google Scholar]
- 71.Brooke J. SUS: a “quick and dirty” usability scale. In: Jordan P, Thomas B, Weerdmeester B, McClelland A, editors. Usability Evaluation in Industry. Taylor & Francis; London: 1996. p. 194. [Google Scholar]
- 72.Iqbal SU, Rogers W, Selim A, et al. The veterans RAND 12 item health survey (VR-12): what it is and how it is used. [Accessed January 16th, 2014]; http://www.herc.research.va.gov/resources/faq_a07.asp.
- 73.DuBenske LL, Burke Beckjord E, Hawkins RP, Gustafson DH. Psychometric evaluation of the Health Information Orientation Scale: a brief measure for assessing health information engagement and apprehension. J Health Psychol. 2009;14:721–730. doi: 10.1177/1359105309338892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hughes C, Lerman C, Schwartz M, et al. All in the family: evaluation of the process and content of sisters' communication about BRCA1 and BRCA2 genetic test results. Am J Med Genet. 2002;107:143–150. doi: 10.1002/ajmg.10110. [DOI] [PubMed] [Google Scholar]
- 75.O'Connor AM. Validation of a decisional conflict scale. Med Decis Making. 1995;15:25–30. doi: 10.1177/0272989X9501500105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.