Figure 1.
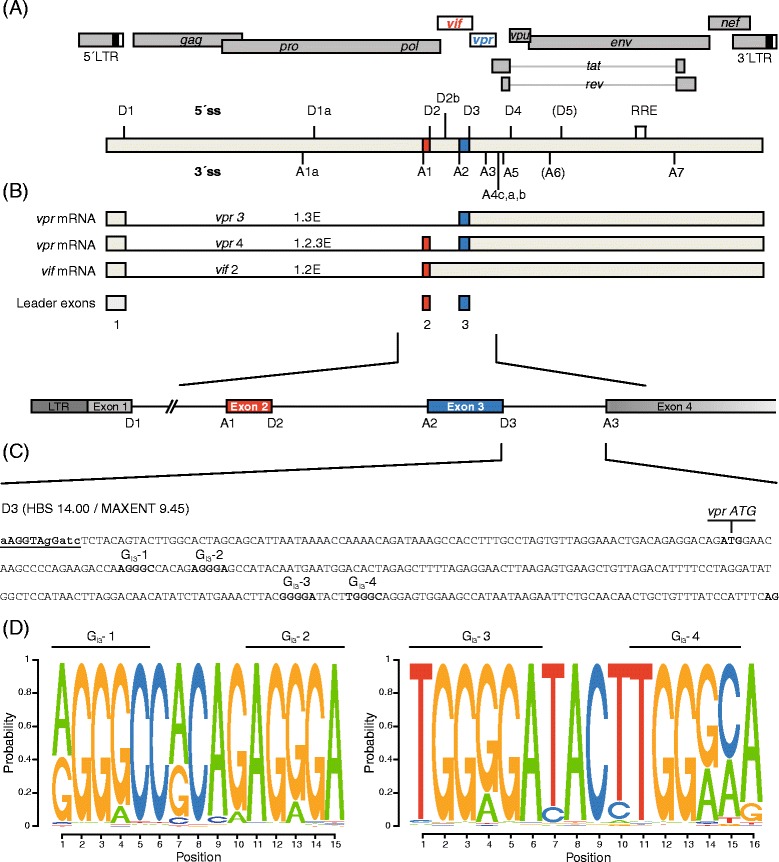
Schematic drawing of the HIV-1 NL4-3 genome. (A) The diagram illustrates the HIV-1 provirus genome including locations of open reading frames (ORFs), long terminal repeats (LTRs), 5′ and 3′ splice sites (ss), exons, introns, and the Rev response element (RRE). Vif and vpr exons and ORFs are highlighted in red and blue, respectively. The RRE is indicated by an open box. (B) Vif and vpr mRNA are formed primarily by splicing of 5′ss D1 to 3′ss A1 and A2, respectively. The noncoding leader exons 2 and 3 are included and AUG-containing introns are retained. (C) Sequence of intron 3 in the proviral HIV-1 genome including the locations of the vpr AUG and G runs GI3-1 to GI3-4. The intrinsic strength of 5′ss D3 is indicated (HBS, MAXENT). (D) Sequence logos generated from a sequence alignment of the HXB2 regions from nt 5581 - nt 5597 and nt 5708 - nt 5723, respectively, flanking the four G runs GI3-1 to GI3-4. Sequences were obtained from the Los Alamos HIV Sequence Database (http://www.hiv.lanl.gov). The relative size of the letters reflects the relative frequency of the nucleotides at the position in the alignment.
