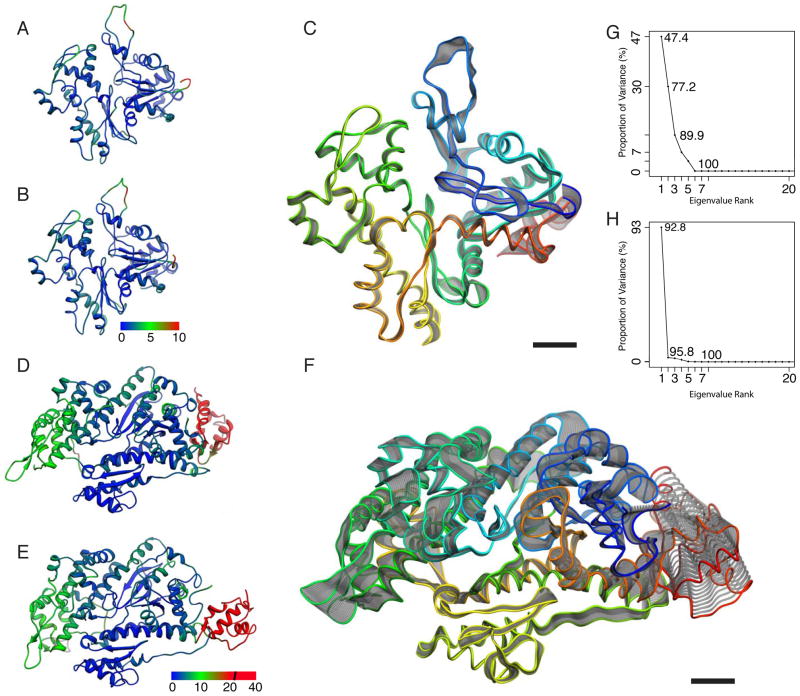
Figure 3. Analysis of conformational changes induced by complex formation.
(A–C) Analysis of the variability of the F-actin models between the undecorated and with myoE and tropomyosin decorated state. (A–B) Atomic models of an undecorated and decorated (group 3) F-actin subunit, respectively. Models are color coded by Cα RMSD values ranging from 0 Å (blue) to 10 Å (red). Major differences are found in carboxy terminal region of the DNase I binding loop (residues 39–52) and the amino terminal region (residues 1–5). (C) Visualization of the first eigenvector obtained after PCA as a trajectory of standard deviation scaled displacements from the average structure. No clear pattern of displacements is discernable. Actin is colored with a rainbow gradient from blue (amino terminus) to red (carboxy terminus). (D–F) Analysis of the variability of the myoE models between pre-power stroke and rigor state. (D–E) Atomic models of pre-power stroke (PDB ID: 1LKX chain C) and rigor state (group 3 myosin) color coded by Cα RMSD values ranging from 0 Å (blue) to 20 Å (red). In the converter domain, displacements ranged up to 40 Å. The second highest displacement locates to the UD50. (F) Visualization of the first eigenvector for myoE. While the LD50, especially the helix-loop-helix motif, is almost invariable, clear changes are seen in all other domains indicating closure of the 50-kDa-cleft and rotation of the converter domain. Myosin is colored with a rainbow gradient from blue (amino terminus) to red (carboxy terminus). (G–H) Eigenvalue spectra for actin PCA and myoE PCA, respectively. While changes upon complex formation cannot easily be accounted for in the case of actin, one eigenvector is enough to account for over 92 % of observed variance in the case of myosin. Scale bar, 1 nm.