Figure 1.
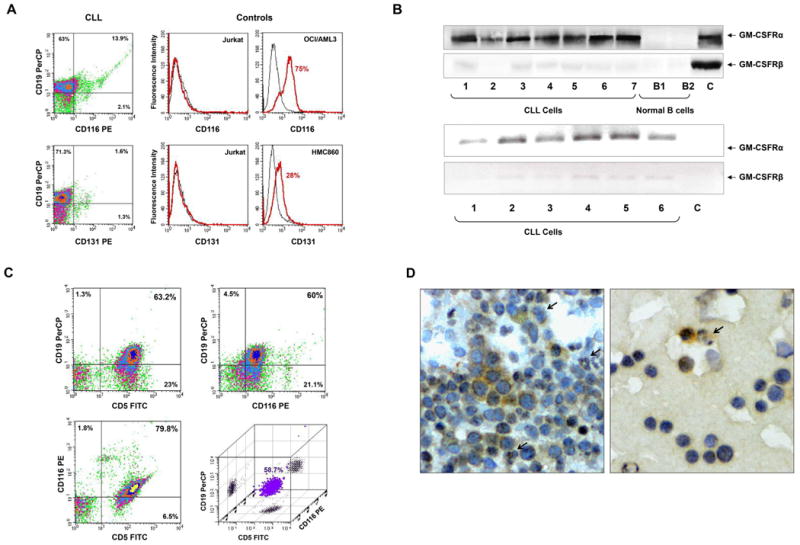
Chronic lymphocytic leukemia (CLL) cells express GM-CSFRα. (A) Flow cytometry analysis of peripheral blood (PB) CLL cells. Low-density cells from a patient with CLL were used. As depicted in the left panel, CLL CD19+ cells express CD116 but not CD131. PerCP is the isotype for CD19 and PE is the isotype controls for CD116 and CD131 are not shown. The positive and negative controls for CD116 and CD131 antibodies are depicted in the right panel. Jurkat cells were used as a negative control, and the myeloid lines OCI/AML3 and HMC860 were used as positive controls. (B) Upper panel: PCR analysis of low-density cells from different CLL patients and negative and normal B cells. As shown, GM-CSFRα was readily detected in all samples whereas very weak signals of GM-CSFRβ transcripts were detected by a 50 cycle touchdown PCR program. Lower panel: Western blot analysis of protein obtained from PB low-density cells of 7 patients with CLL and CD19+ B-lymphocytes from 2 healthy volunteers. As shown, GM-CSFRα, but not GM-CSFRβ, is detected in all CLL samples but not in normal B-cells. (C) Three-color flow cytometry analysis of PB low-density CLL cells. As shown, 63.2% of the cells co-expressed CD5 and CD19, 79.8% co-expressed CD5 and CD116, 60% co-expressed CD19 and CD116, and 58.7% co-expressed CD5, CD19 and CD116 (PerCP, peridinin-chlorophyll protein). (D) Immunohistochemistry staining with anti-GM-CSFRα antibodies of CLL BM clot (left panel) and smear (right panel). Several mononuclear cells, some of which exhibit typical CLL cell morphological features, stained positively for GM-CSFRα, whereas other lymphocytes did not. Both cytoplasmic and nuclear speckles are seen. Arrows depict positively stained granulocytes (positive control).
