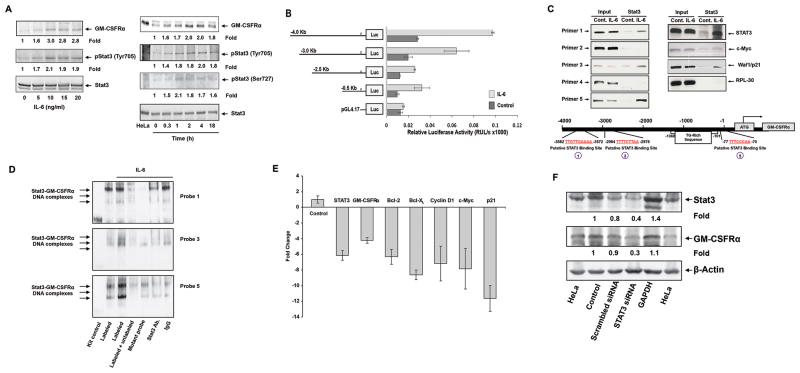
Figure 3.
IL-6-activated STAT3 activates the GM-CSFRα promoter and induces GM-CSFRα protein production in multiple myeloma cell line MM1 cells. (A) IL-6 phosphorylates STAT3 and upregulates GM-CSFRα protein levels in MM1 cells. Left panel: MM1 cells were incubated for 2 hours with increasing concentrations of IL-6. As determined by Western immunoblotting, IL-6 induced STAT3 tyrosine phosphorylation and upregulated GM-CSFRα protein levels in a dose-dependent manner. Right panel: MM1 cells were incubated with 20 ng/ml of IL-6 and harvested at different time points for further analysis. IL-6 induced STAT3 phosphorylation and upregulated GM-CSFRα protein levels in a time-dependent manner. (B) IL-6 induces GM-CSFRα promoter activity. A schematic diagram of the GM-CSFRα reporter fragments that were transfected into MM1 cells and maintained in the absence or presence of 20 ng/ml of IL-6 is presented on the left panel. The right panel depicts the mean ± standard deviation of the relative luciferase activity measured in 4 different experiments in which transfection efficiency of the truncated GM-CSFRα promoter constructs into MM1 cells was at least 45%. IL-6 induced luciferase activity in MM1 transfected with promoter fragments -4.0, -3.0, -2.5, and -0.5 Kb. Fragments -2.5 and 0.5 Kb yielded a similar IL-6-induced luciferase activity. IL-6 did not significantly increase luciferase activity in PGL4.17. (C) STAT3 binds to the promoter of GM-CSFRα and other STAT3-regulated genes. ChIP demonstrates that anti-STAT3 antibodies immunoprecipitated GM-CSFRα (left panel) and the STAT3-regulated genes STAT3, c-Myc, and WAF1/p21 (right panel). These genes were detected in MM1 cell nuclear extracts (Input) as well as in nuclear protein immunoprecipitated with anti-STAT3 antibodies in IL-6-stimulated MM1 cells, suggesting that STAT3 binds to known STAT3-regulated gene promoters and to the GM-CSFRα promoter. As shown in the left panel, binding of STAT3 to the GM-CSFRα promoter is detected by primers 1, 3, and 5, but not 2 and 4. The schematic diagram in the lower panel depicts the 3 active (1, 3, and 5) STAT3 binding sites. (D) EMSA results, using biotin-labeled and unlabeled DNA probes 1, 3, and 5 (Supplementary Table 2), show that IL-6-stimulated MM1 cell nuclear protein binds to the biotinylated DNA fragments 1, 3, and 5 and that the addition of the corresponding cold (unlabeled) DNA fragments or anti-STAT3 antibodies (but not IgG) attenuates the binding. The binding of IL-6-stimulated MM1 cell nuclear protein to biotinylated mutated DNA fragments 1, 3, and 5 (Table 2) is diminished or significantly reduced. (E) qRT-PCR demonstrates that STAT3-siRNA downregulates the mRNA levels of GM-CSFRα, and the STAT3-regulated genes STAT3, ROR1, c-Myc, cyclin D1, and p21. S18 mRNA (control) was not affected. (F) Western blot analysis shows that STAT3-siRNA, but not scrambled-siRNA or GAPDH, downregulates the protein levels of STAT3 and GM-CSFRα.