Figure 9.
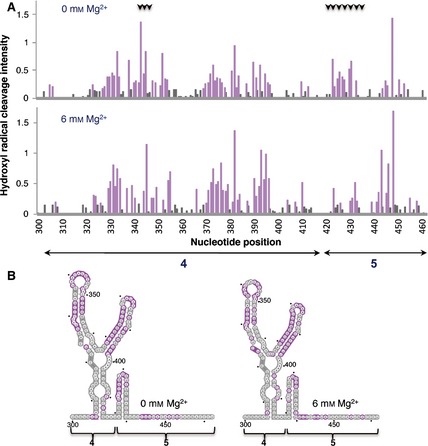
Hydroxyl radical probing of IRES domains 4 and 5. (A) Histograms of cleavage intensity versus nucleotide position in the absence or presence of Mg2+ determined using in situ‐generated hydroxyl radicals. Cleavages were determined by primer extension using fluorescent‐labeled primers and capillary electrophoresis. Gray bars indicate solvent‐inaccessible nucleotides in the folded RNA (intensities < half the mean), and solvent‐accessible residues are shown in purple. Black arrows at the top indicate positions whose accessibility decreases upon increasing concentration of Mg2+ in the folding buffer. (B) Predicted secondary RNA structure of domains 4 and 5 of the IRES imposing SHAPE constraints. Solvent‐inaccessible nucleotides are shown in gray; solvent‐accessible nucleotides are shown in purple.
