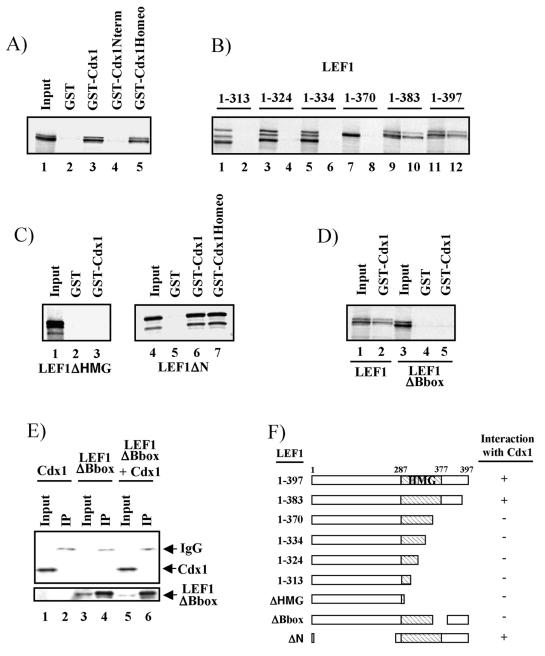
FIG. 4.
Cdx1 interacts directly with LEF1. GST-Cdx1 and 35S-labeled LEF1 constructs were generated and assessed for interaction as described in Materials and Methods. (A) Both full-length Cdx1 (GST-Cdx1) and the Cdx1 homeodomain (GST-Cdx1Homeo) interact with LEF1 (lanes 3 and 5), whereas Cdx1 N-terminal sequences do not (lane 4). (B) Deletion of sequences N-terminal to position 383 of LEF1 abolished interaction with Cdx1 (compare lane 10 to lane 8; odd-numbered lanes are 20% input of the various 35S-labeled LEF1 constructs, and even-numbered lanes are from the GST-Cdx1 pulldown). (C) Deletion of the LEF1 HMG box (lane 3) abolishes interaction with Cdx1, whereas loss of N-terminal LEF1 residues has no effect on interaction with either full-length Cdx1 (lane 6) or the homeodomain sequences of Cdx1 (lane 7). (D and E) Deletion of the LEF1 B box abolishes interaction with Cdx1 in vitro (D, compare lane 5 to lane 2) and in vivo (E, lane 6). (F) Schematic representation of LEF1-Cdx1 interactions. Coimmunoprecipitations (panel E) were performed as in Fig. 2 except the ΔBbox mutant was used instead of full-length LEF1.