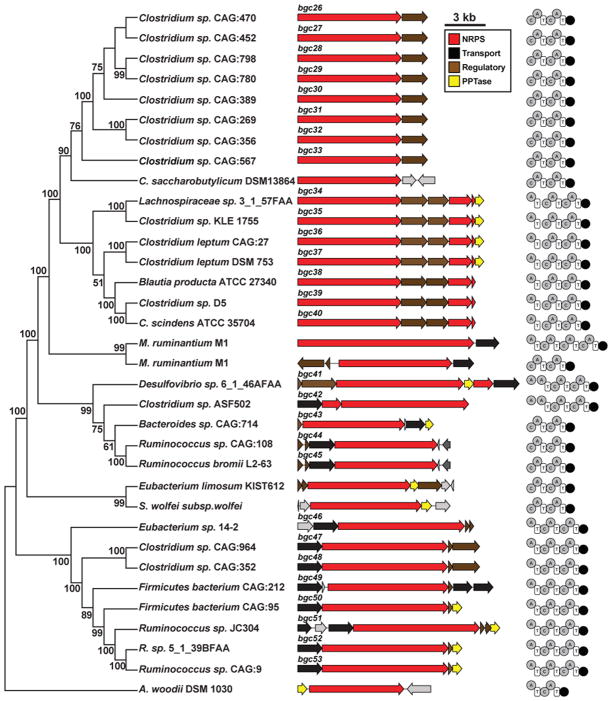
Figure 3. An abundant family of NRPS BGCs is found exclusively in gut isolates and stool metagenomes.
Left, a phylogenetic tree (Maximum Parsimony, MEGA5) based on the main NRPS gene of 28 clusters from human bacterial gut isolates, two from bovine rumen archaeal isolates, and four from environmental isolates (see Experimental Procedures). The numbers next to the branches represent the percentage of replicate trees in which this topology was reached in a bootstrap test of 1000 replicates. Middle, schematic of each BGC (see also Figure S3 for a full heat map showing the prevalence and abundance of each member of this NRPS family in HMP stool samples). Right, domain organization of the NRPS genes of each cluster (A, adenylation domain; C, condensation domain; T, thiolation domain; R, terminal reductase domain).