Figure 6. Characterization of a new antibiotic, lactocillin, from a human vaginal isolate.
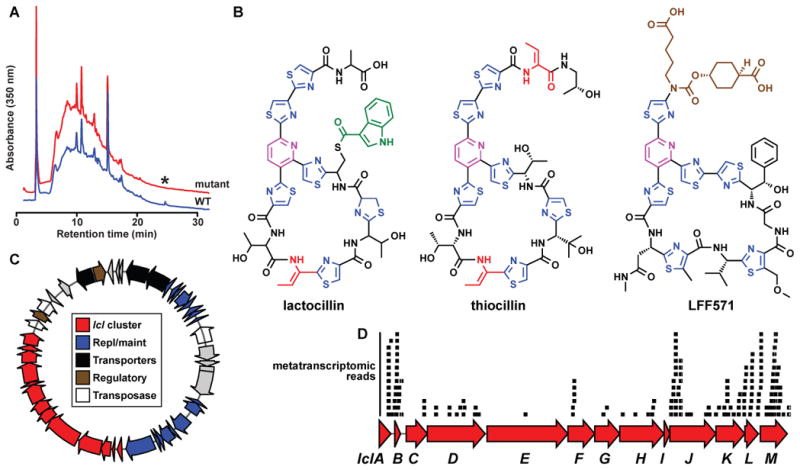
A) HPLC analysis of organic extracts of cell pellets from wild-type (red) and lclD insertional mutant (blue) strains of L. gasseri JV-V03, monitored at 350 nm. An asterisk indicates the HPLC peak corresponding to lactocillin, B) Planar structure of lactocillin (see Experimental Procedures and see also Figure S6 and Supplemental Data File 2 for details about its purification and structural elucidation), the Bacillus cereus antibiotic thiocillin, and the clinical candidate LFF571 (Phase II, Novartis). Note the structural similarities among the three thiopeptides. C) Plasmid harboring the lactocillin BGC (see Experimental Procedures for details of the experimental closure of the circular plasmid). The lactocillin gene cluster (red) occupies ~30% of the plasmid; other elements on the plasmid include plasmid replication and maintenance genes (blue), transporters (black), transcription regulators (brown) and transposases and phage integrases (white). D) Raw oral metatranscriptomic reads were recruited to the lcl cluster using blastn and aligned using Geneious. Each black bar represents one read. Note that the precursor peptide lclB is amongst the most deeply covered genes in the lcl cluster, as anticipated for a RiPP pathway (see Experimental Procedures and see also Figure S7 for metatranscriptomic analysis of the whole lcl plasmid, and of bgc65).
