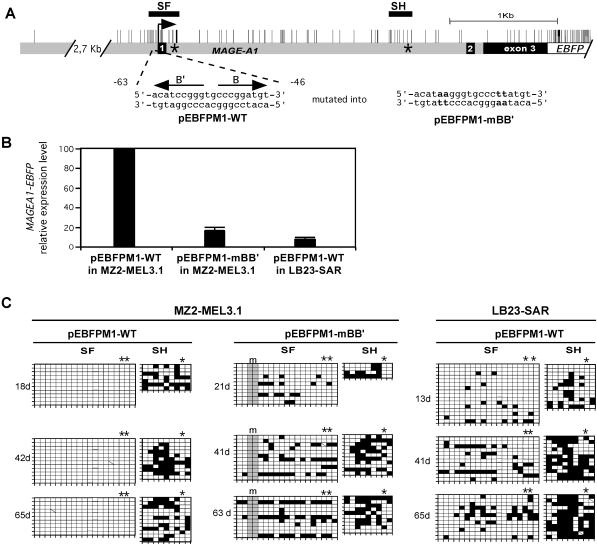
FIG. 5.
Protection of the MAGE-A1 5′ region from remethylation depends on promoter activity. (A) The MAGE-A1 gene fragment cloned in front of the EBFP transcription unit in the pEBFPM1-WT and pEBFPM1-mBB′ constructs is represented as in Fig. 1. Positions of the SF and SH segments are indicated by horizontal bars. Asterisks indicate the positions where short oligonucleotide tags were introduced. The wild-type and mutated MAGE-A1 promoter sequences (present in pEBFPM1-WT and pEBFPM1-mBB′, respectively) are given, with arrows indicating the positions and orientations of B and B′ regulatory elements. (B) MZ2-MEL3.1 and LB23-SAR cells were stably transfected with unmethylated pEBFPM1-WT or pEBFPM1-mBB′ plasmid DNA. Quantitative real-time RT-PCR was used to measure the relative MAGE-A1/EBFP expression levels in the transfectants at the earliest time point after transfection. MZ2-MEL3.1 transfected with pEBFPM1-WT was taken as the 100% reference. Values are the means for three separate RT-PCR experiments. (C) Methylation patterns in the SF and SH segments of the MAGE-A1 transgenes are represented by the grids, as in Fig. 1. Sequences deriving from the exogenous MAGE-A1 transgenes could be distinguished from those deriving from the endogenous MAGE-A1 gene by the presence of sequence tags. Asterisks above the grids indicate CpG sites deriving from these tag sequences. The mutations in the B and B′ promoter elements (m) resulted in the loss of two CpG sites, as indicated by the shading of the corresponding columns.