Abstract
The PER3 gene is one of the clock genes, which function in the core mammalian molecular circadian system. A variable number of tandem repeats (VNTR) locus in the 18th exon of this gene has been strongly associated to circadian rhythm phenotypes and sleep organization in humans, but it has not been identified in other mammals except primates. To better understand the evolution and the placement of the PER3 VNTR in a phylogenetical context, the present study enlarges the investigation about the presence and the structure of this variable region in a large sample of primate species and other mammals. The analysis of the results has revealed that the PER3 VNTR occurs exclusively in simiiforme primates and that the number of copies of the primitive unit ranges from 2 to 11 across different primate species. Two transposable elements surrounding the 18th exon of PER3 were found in primates with published genome sequences, including the tarsiiforme Tarsius syrichta, which lacks the VNTR. These results suggest that this VNTR may have evolved in a common ancestor of the simiiforme branch and that the evolutionary copy number differentiation of this VNTR may be associated with primate simiiformes sleep and circadian phenotype patterns.
Introduction
The light/dark cycle that arises from the geospatial relationship between the Earth and the Sun has been shown to be the strongest environmental clue, or zeitgeber, for the entrainment of circadian rhythms in many organisms, including human beings [1], [2]. Virtually all physiological processes, such as hormone secretion, sleep, and body temperature which oscillate with circadian periodicity, are regulated by the light/dark signal [3]. The processing of this signal is associated with the expression of a group of genes, collectively known as clock genes, which are directly involved in the regulation and maintenance of the circadian rhythms [4], [5]. Due to the fact that variations in these genes, such as polymorphisms and mutations, are associated with aberrant or differential expression of circadian behaviors (reviewed in [6]), the clock genes are strong candidates for natural selection associated with the light/dark cycle signal.
Period circadian clock 3 (PER3) is one of the clock genes engaged in the core mammalian molecular circadian system [7], [8]. In humans, this gene is located on the short arm of chromosome 1 (1p36) [9], and its 18th exon contains a polymorphic variable number of tandem repeats (VNTR) composed of a 54-bp motif that is repeated either four or five times [10]. This length polymorphism in the PER3 gene has been described in few primate species, such as chimpanzees, gorillas, gibbons, langurs, marmosets, and orangutans, but not in non-primate mammals, such as rats, dogs, and mice [11]. Several reports have shown associations between this VNTR and human circadian rhythm phenotypes, including morning/evening preferences, delayed sleep phase disorder (DSPD), and homeostatic regulation of sleep [10], [12]–[16], highlighting an important link between genetics, the sleep/wake cycle and adaptation to the light/dark cycle.
Due to the importance of the PER3 VNTR for regulation of sleep and circadian rhythms in humans, it is a matter of interest to investigate the hypothesis that this genomic structure is exclusive to primates. This would allow rationalizing whether this VNTR is associated to some special feature on the primate circadian system. Since, with very few expections, Simiiforme primates are essentially diurnal or cathemeral animals [17], [18] and Prosimians are almost all nocturnal, it would be pertinent to ask whether this VNTR is associated with activity phase allocation during the day or the night. The present study aimed to analyze and compare the PER3 VNTR regions of several primate species and other non-primate mammals.
Materials and Methods
Biological Material from Primate Species
Blood samples were collected from 129 individuals belonging to 13 New World primate species (Table 1) maintained at the Tufted Capuchin Procreation Center of Universidade Estadual Paulista (Araçatuba campus, Brazil), at the Zoological Park of São Paulo, at Centro Nacional de Primatas (CENP, Ananindeua, Pará, Brazil), at the Marmoset Vivarium of Universidade Federal de São Paulo (São Paulo campus), and at Centro de Primatas of Universidade Federal do Rio Grande do Norte.
Table 1. Primate species analyzed in the present study.
| Common name | Species name | Family | Order | IndividualsAnalyzed | GenBankaccession n# | Genome coverage |
| Grivet | Chlorocebus aethiops | Cercopithecidae | Primates | 2 | KC146097 | * |
| Goeldi’s marmoset | Callimico goeldii | Cebidae | Primates | 2 | KC146101 | * |
| Common marmoset | Callithrix (Callithrix) jacchus | Cebidae | Primates | 80 | KC146106 | 6X (Mar 2009) |
| White-headed marmoset | Callithrix (Callithrix) geoffroyi | Cebidae | Primates | 1 | KC146096 | * |
| Red-handed tamarin | Saguinus midas | Cebidae | Primates | 1 | KC146100 | * |
| Emperor tamarin | Saguinus imperator | Cebidae | Primates | 1 | KC146098 | * |
| Brown-mantled tamarin | Saguinus fuscicollis | Cebidae | Primates | 1 | KF408263/KF408264 | * |
| Tufted capuchin | Cebus apella | Cebidae | Primates | 33 | KC146102 | * |
| Common squirrel monkey | Saimiri sciureus | Cebidae | Primates | 1 | KC146099 | * |
| Kuhl’s owl monkey | Aotus azarae infulatus | Aotidae | Primates | 2 | KC146095 | * |
| Black howler | Alouatta caraya | Atelidae | Primates | 2 | KC146105 | * |
| Red-faced spider monkey | Ateles paniscus | Atelidae | Primates | 2 | KC146104 | * |
| Brown woolly monkey | Lagothrix lagotricha | Atelidae | Primates | 1 | KC146103 | * |
*Genomic assembly information not available.
Classification according to “Wilson & Reeder’s Mammal Species of the World, 3rd Edition”, available at the Taxonomic Browser of the Smithsonian National Museum of Natural History <http://www.vertebrates.si.edu/msw/mswcfapp/msw/index.cfm>.
This study was approved by the Local Committee for Ethics in Research of Universidade Federal de São Paulo (UNIFESP/EPM 0136/05) and includes authorization for sample collections at all animal facilities located inside the universities and within the zoological park. Blood collection concerning wild animals was authorized by the Brazilian Institute of Environment and Renewable Natural Resources (IBAMA) under license #16198. IBAMA, through Deliberation #40/2003 of the Ministry of Environment and the Genetic Heritage Management Council, is the institution responsible for issuing permits for access and delivery of genetic heritage for research activities in the area of biology throughout the Brazilian territory.
All animals were housed under natural conditions of temperature and humidity in a light-dark cycle. The cages were constructed in brick and wire mesh measuring 0.9×2.0×1.8 m, equipped with wood, rope, perches, basket, concrete platforms, cooler box and nest platforms. Water was available ad libitum, and food was provided twice a day: in the morning hours between 7∶00 and 9∶00 am, and in the afternoon between 1∶00 and 3∶00 pm. The feed consisted of seasonal fruits and a rich-in-protein preparation that supplemented the diet. Candies were also sporadically offered to animals. At the end of blood sampling the animals were not euthanized. During blood collection, the animals were immobilized by suitable equipment. All animals were in excellent health. After collection the animals received a bounty of candy. The experimental procedures were in accordance with the Guidelines for the Care and Use of Mammals in Neuroscience and Behavioral Research of the National Research Council.
Extraction, gene amplification, and sequencing
Blood samples were collected into PAXgene Blood RNA vacutainer tubes (PreAnalytiX, Hombrechtikon, Switzerland), and frozen at –20°C. Total RNA or genomic DNA (gDNA) were extracted from the animals’ white blood cells using the PAXgene 96 Blood RNA Kit (QIAGEN, Hilden, Germany). The target cDNA or gDNA regions were amplified by PCR using the primers described by Jenkins and colleagues (5′ AGCAGYTCACCSTTRCAGTT 3′ and 5′ GGYACCTGGTATGTCATGAGAA 3′) [11] or the following pair of primers: 5′ GACTAACAGGTGGGTGGCA 3′ and 5′ CAGAACTTTTTGGGGTGAC 3′. The PCR amplicons were sequenced, and the sequences obtained were submitted to NCBI GenBank (Table 1).
In silico analysis an comparisons of the VNTR region sequence among primate and non-primate species
Multiple alignments of the PER3 nucleotide sequences obtained were performed using the ClustalW algorithm [19]. The alignments were then visually examined to ascertain the presence of the VNTR and the number of repeats in each primate species. In addition, the PER3 protein sequences from several mammalian species, including prosimians (Table 2), were obtained from the Ensembl Genome Browser Database (http://www.ensembl.org) and were examined for the presence of the target VNTR by aligning the human PER3 protein sequence and the orthologous region from the selected mammals. In silico searches for repetitive elements surrounding the VNTR were conducted using the RepeatMasker 3.1.8 online software (http://www.repeatmasker.org).
Table 2. Non-simiiforme primates and non-primate mammal species analyzed in the present study.
| Common name | Species name | Family | Order | Ensembl Gene ID | Genome coverage |
| Gray short-tailed opossum | Monodelphis domestica | Didelphidae | Didelphimorphia | ENSMODG00000005802 | 7.33X (Out 2006) |
| Tammar wallaby | Macropus (N.) eugenii | Macropodidae | Diprotodontia | ENSMEUG00000010189 | 2X (Nov 2009) |
| Rock hyrax | Procavia capensis | Procaviidae | Hyracoidea | ENSPCAG00000004848 | 2.19 (Jul 2008) |
| African bush elephant | Loxodonta africana | Elephantidae | Proboscidea | ENSLAFG00000000581 | 7X (Jul 2009) |
| Nine-banded armadillo | Dasypus novemcinctus | Dasypodidae | Cingulata | ENSDNOG00000009963 | 2X (May 2009) |
| Hoffmann’s two-toed sloth | Choloepus hoffmanni | Megalonychidae | Pilosa | ENSCHOG00000007396 | 2.05X (Sep 2008) |
| Northern treeshrew | Tupaia belangeri | Tupaiidae | Scandentia | ENSTBEG00000003546 | 2X (Jul 2006) |
| Gray mouse lemur | Microcebus murinus | Cheirogaleidae | Primates | ENSMICG00000008437 | 1.93X (Jun 2007) |
| Northern greater galago | Otolemur garnettii | Galagidae | Primates | ENSOGAG00000009800 | 1.5X (Mar 2011) |
| Philippine tarsier | Tarsius syrichta | Tarsiidae | Primates | ENSTSYG00000007736 | 1.82X (Jul 2008) |
| Guinea pig | Cavia porcellus | Caviidae | Rodentia | ENSCPOG00000022169 | 6.79X (Mar 2008) |
| Ord’s kangaroo rat | Dipodomys ordii | Heteromyidae | Rodentia | ENSDORG00000015158 | 1.85X (Jul 2008) |
| Thirteen-lined ground squirrel | Spermophilus (I.) tridecemlineatus | Sciuridae | Rodentia | ENSSTOG00000014007 | 2X (Nov 2011) |
| Large flying fox | Pteropus vampyrus | Pteropodidae | Chiroptera | ENSPVAG00000005395 | 2.63X (Jul 2008) |
| Little brown myotis | Myotis lucifugus | Vespertilionidae | Chiroptera | ENSMLUG00000015940 | 7X (Sep 2010) |
| Domestic cat | Felis catus | Felidae | Carnivora | ENSFCAG00000004371 | |
| Horse | Equus caballus | Equidae | Perissodactyla | ENSECAG00000018928 | 6.79X (Sep 2009) |
| Bovine | Bos taurus | Bovidae | Artiodactyla | ENSBTAG00000014604 | |
| Bottlenose dolphin | Tursiops truncatus | Delphinidae | Cetacea | ENSTTRG00000010402 | 2.59X (Jul 2008) |
Classification according to “Wilson & Reeder’s Mammal Species of the World, 3rd Edition”, available at the Taxonomic Browser of the Smithsonian National Museum of Natural History <http://www.vertebrates.si.edu/msw/mswcfapp/msw/index.cfm>.
Results
The DNA analysis of the PER3 VNTR region belonging to the New World primates revealed that all of the studied individuals carried this locus in their genomes, although the number of repeats varied among the different species (Figure 1 and Figure S1). The tufted capuchins and Kuhl’s owl monkeys analyzed had only two repeats. The brown woolly monkey, the two grivets and the red-handed tamarin had three repeats, whereas the two black howlers, the two red-faced spider monkeys and the common squirrel monkey had four. The emperor tamarin had five repeats, the two Goeldi’s marmosets had six, and the white-headed marmoset had seven (Figure 1).
Figure 1. Schematic representation of the alignment of the PER3 VNTR regions from several primate species.
Each rectangle represents one repeat of the VNTR. *Sequences obtained from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). **Sequences obtained from Jenkins et al. (2005).
The analysis of the PER3 protein sequences from several mammalian species revealed that the amino acid sequence in this region differed both in size and sequence among species (Figure 2). No full repeats of the 18 amino acids of this VNTR were found among the prosimians and non-primate mammals and, although some of the non-primate mammalian sequences aligned with one of the repeat units of the human VNTR, the alignment score was low. Only a few species exhibited scattered amino acid identity to the amino acids present in a human unit of the VNTR sequence. Thus, the non-primate mammals examined in the present study had neither repeats of this VNTR nor a single sequence identical to a human unit of the 18 amino acids. We would like to emphasize that the in silico analysis of the PER3 VNTR region of three species of prosimian primates (Tarsius syrichta, Otolemur garnettii, and Microcebus murinus) also revealed an absence of repeats (Figure 2).
Figure 2. Alignment of the PER3 protein sequences from diverse mammal species, in the region corresponding to the human PER3 VNTR.
The sequences were obtained from the Ensembl Genome Browser Database (http://www.ensembl.org). Dots (.) indicate identity with human sequence. Dashes (–) indicate gaps. X’s indicate unknown or unspecified aminoacids.
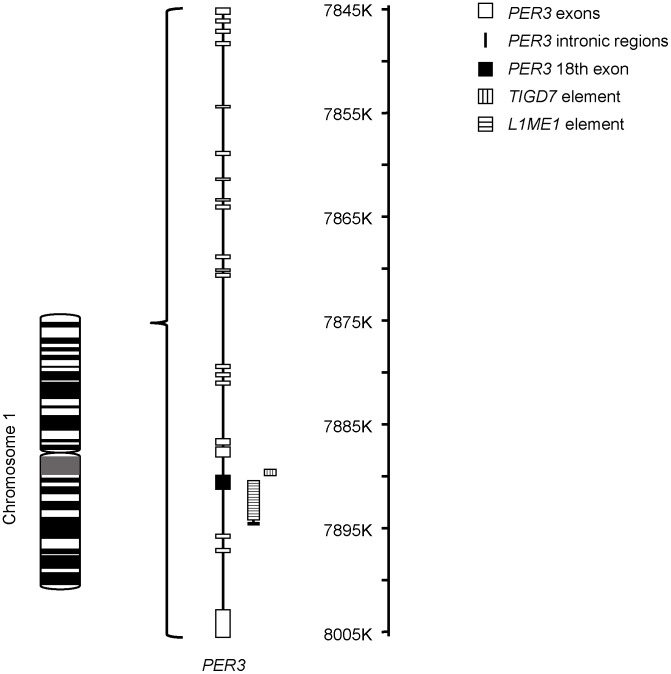
In addition, the bioinformatics analysis showed that the PER3 VNTR is surrounded by transposable elements. The DNA transposon tigger transposable element derived 7 (TIGD7) is inserted upstream of the VNTR at the end of the intron and a few nucleotides before the beginning of the first repeat. Downstream of the VNTR, a LINE-1 element (L1ME1) is inserted near the end of the last repeat (Figure 3). Interestingly enough, although the prosimian Tarsius syrichta contains these two transposable elements in this region of the gene, such species does not have the VNTR. Moreover, the non-primate mammals analyzed did not exhibit transposable elements nor VNTR.
Figure 3. Representation of the PER3 human gene and the repetitive sequences associated to it.
The black square represents the 18th exon of PER3, where the VNTR is placed. Adapted from NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/projects/mapview).
Discussion
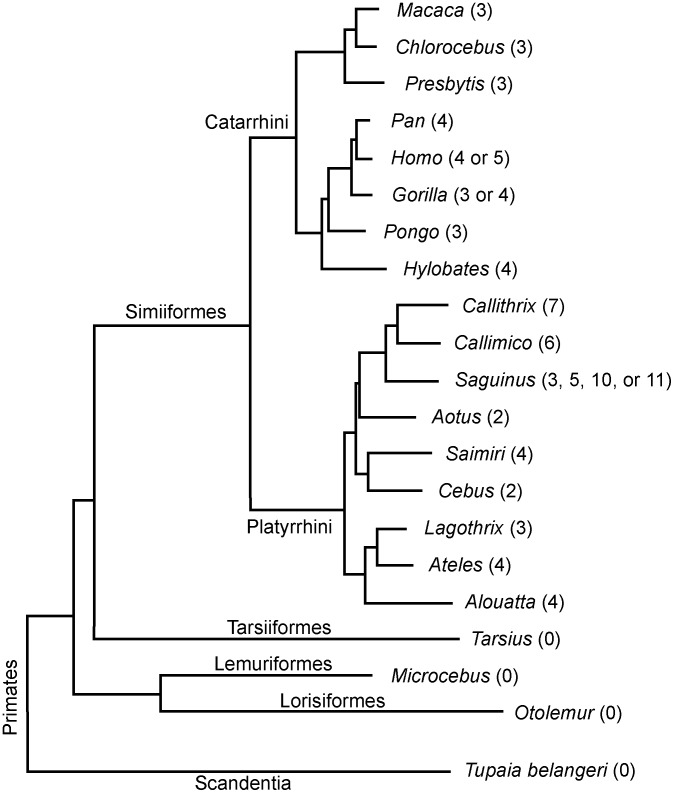
The present study demonstrates that the VNTR in the PER3 gene is a genomic structure present in all Simiiforme primates studied, although the number of repeats is different, depending on the species. We have found that the number of repeats range from 2 to 11 (Figure 1). Observation of these results indicates that other similar primates, not studied here, may also carry the VNTR, thus indicating that this structure is present exclusively in primates [11]. Platyrrhini (New World monkeys) exhibits greater variability. For example, the species belonging to genus Aotus (the only established nocturnal simiiforme primate) exhibit only two repeats, whereas genus Saguinus carries up to 11 repeats.
In silico analysis of the flanking regions concerning the 18th exon of PER3 revealed that both introns adjacent to the VNTR contain transposable elements (Figure 3). The sequences of the two transposable elements surrounding the PER3 VNTR region are highly similar in all the primates analyzed, including the prosimian Tarsius syrichta, and are absent in non-primate mammals. This finding suggests that these elements were present in the common ancestor of Tarsiiformes and Simiiformes, which probably lived during the Eocene Period [20], [21], and that the insertion of the transposable elements occurred prior to the appearance of the PER3 VNTR. Moreover, the absence of these sequences in non-primate mammals indicates that their insertion into these loci was a primate lineage-specific event.
Transposable elements are known to produce genomic instability, rearrangements, genetic innovation [22]–[28], and are associated with the generation of tandem repeats [29]. Therefore, one might tentatively propose that the primitive insertion of these transposable elements in the primate lineage caused the emergence of the VNTR in the PER3 gene in this Order.
The results of the present study, combined with data generated in a previous report [11], enable a reasonable reconstruction of the primate PER3 VNTR evolution (Figure 4). The interpretation of these results largely follows the explanation provided by Jenkins and colleagues [11] who suggest that the VNTR was derived from a single ancestral unit in the common ancestor of primates, carnivores, and rodents. However, data from the additional species analyzed herein and the additional finding of transposable elements flanking the VNTR region suggest that, in fact, the VNTR derived from a single copy that was present in the common ancestor of Simiiformes, and that a duplication event most likely occurred in primates before Catarrhini and Platyrrhini diverged, approximately 43.5 million years ago [20], [21]. The repeat thus created was then subject to a variable number of posterior duplications and, perhaps, deletions.
Figure 4. Evolutionary relationships among the analyzed primate genera, based on the phylogeny proposed by Perelman et al. (2011).
Between brackets, the number of repetitions comprising the PER3 VNTR for each genus.
Tandem repeats exhibit mutation rates much higher than those of other genomic regions. When they are located inside genes and regulatory regions, tandem repeats may influence the properties or functions of these genes, such as binding sites, chromatin structure, and, ultimately, gene transcription [30]. A VNTR that is located within a coding region may affect the stability and/or activity of the gene product [31], [32]. Although molecular mechanisms underlying gene functional changes mediated by tandem repeats are generally poorly understood, tandem repeat domains in proteins are usually involved in protein-protein interactions and fine-tuning protein conformation [33]. Nevertheless, it is unlikely that variations in tandem repeats produce completely new characteristics; rather, they allow the fine-tuning of specific phenotypes, including those related to behavior, physiology, and morphology, due to the wide range of allelic variation [34], thus switching from most night to most day activity patterns; in the case of primates, would fit with the fine-tuning phenotype proposal. A clear documented case for this phenomenon, for instance, is that variations in the number of repeats of a hexanucleotide in exon 5 of the Period gene influence the mechanism of temperature compensation in Drosophila melanogaster [35], [36].
The uniqueness of the PER3 VNTR in primates and its strong association with circadian phenotypes and homeostatic sleep regulation in humans indicate that this VNTR is associated with a special feature of the primate circadian system or sleep. When compared with other mammalian species, Simiiforme primates exhibit such special features as consolidated monophasic or biphasic sleep [37] and a diurnal activity pattern. The owl monkeys from the genus Aotus are an exception between the Simiiformes [38], and the Prosimians, more primitive primates, are mostly nocturnal. Interestingly, the present study showed that the analyzed Aotus species possesses only two repeats of the PER3 VNTR, and the Prosimian species, none.
It is generally assumed that the ancestor of mammals - and of primates in particular - was a nocturnal creature [39]. Although most of the recently evolved mammals are nocturnal, Simiiformes definitively are not. Repetitive sequences may provide a high degree of evolutionary flexibility, allowing adaptive accommodations at a minimal cost to the genetic function. Thus, perhaps the PER3 VNTR is involved as a part of the mechanism that provides evolutionary flexibility that allowed the occupation of different portions of the light/dark niche - in the case of Simiiformes, the lightened portion of the cycle.
While some genomes used for the sequence assembling have high sequencing depths, such as the genomes of human, chimp, mouse and dog, others, such as the gray mouse lemur genome, present only low levels of sequence coverage. Although a complete coverage of the genomes used for the study is obviously preferable, the usage of lower-redundancy genomes may be the only alternative when investigating non-laboratory model species [40]. While low-percent coverage brings some limitations for sequence analysis, utilization of only deeper-covered genomes would substantially reduce the number of targets, crashing the possibility of a phylogenetic approach.
In summary, the present study demonstrates that the PER3 VNTR is a genomic structure found exclusively in the primate PER3, a gene found only among vertebrates. This VNTR is flanked by two transposable elements that are phylogenetically older than the VNTR itself and that may have been involved in the emergence of this structure. The present findings also show that the nocturnal Tarsius does not possess this VNTR and that primates of the genus Aotus, which are nocturnal, have the smallest number of copies of the VNTR of almost all the primates analyzed.
Supporting Information
Alignment of the PER3 VNTR regions from several primate species. Each repetition is represented in a different color. *Sequences obtained from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). **Sequences obtained from Jenkins et al. (2005).
(TIF)
Acknowledgments
We would like to thank Primate National Center, Park Zoo Fundation of São Paulo, Macaco-Prego Center of UNESP Araçatuba campus, Primate Center of Primates of UFRN, and Marmoset Vivarium of UNIFESP São Paulo campus. We would also like to thank Simone C. Maria for providing and allowing the collection of samples and Dr. Horacio de la Iglesia for comments on the manuscript.
Funding Statement
This work was supported by the Foundation for Research Support of the State of Sao Paulo (FAPESP); association incentive fund psychopharmacology (AFIP); center for research innovation and diffusion (CEPID). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Panda S, Hogenescht JB, Kay SA (2002) Circadian rhythms from flies to human. Nature 417: 329–335. [DOI] [PubMed] [Google Scholar]
- 2. Roenneberg T, Kumar CJ, Merrow M (2007) The human circadian clock entrains to sun time. Curr Biol 17: R44–45. [DOI] [PubMed] [Google Scholar]
- 3. Aschoff J (1965) Circadian rhythms in man. Science 148: 1427–1432. [DOI] [PubMed] [Google Scholar]
- 4. Reppert SM, Weaver DR (2001) Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol 63: 647–676. [DOI] [PubMed] [Google Scholar]
- 5. Reppert SM, Weaver DR (2002) Coordination of circadian timing in mammals. Nature 418: 935–941. [DOI] [PubMed] [Google Scholar]
- 6. Albrecht U (2002) Invited review: regulation of mammalian circadian clock genes. J Appl Physiol 92: 1348–1355. [DOI] [PubMed] [Google Scholar]
- 7. Takumi T, Taguchi K, Miyake S, Sakakida Y, Takashima N, et al. (1998) A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J 17: 4753–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zylka MJ, Shearman LP, Weaver DR, Reppert SM (1998) Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20: 1103–1110. [DOI] [PubMed] [Google Scholar]
- 9. Gregory SG, Barlow KF, McLay KE, Kaul R, Swarbreck D, et al. (2006) The DNA sequence and biological annotation of human chromosome 1. Nature 441: 315–321. [DOI] [PubMed] [Google Scholar]
- 10. Ebisawa T, Uchiyama M, Kajimura N, Mishima K, Kamei Y, et al. (2001) Association of structural polymorphisms in the human PERIOD3 gene with delayed sleep phase syndrome. EMBO Rep 2: 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jenkins A, Archer SN, von Schantz M (2005) Expansion during primate radiation of a variable number tandem repeat in the coding region of the circadian clock gene period3. J Biol Rhythms 20: 470–472. [DOI] [PubMed] [Google Scholar]
- 12. Archer SN, Robilliard DL, Skene DJ, Smits M, Williams A, et al. (2003) A lenght polymorphism in the circadian clock gene PER3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep 26: 413–415. [DOI] [PubMed] [Google Scholar]
- 13. Pereira DS, Tufik S, Louzada FM, Benedito-Silva AA, Lopez AR, et al. (2005) Association of the lenght polymorphism in the human PER3 gene with the delayed sleep-phase syndrome: does latitude have an influence upon it? Sleep 28: 29–32. [PubMed] [Google Scholar]
- 14. Viola AU, Archer SN, James LM, Groeger JA, Lo JC, et al. (2007) PER3 polymorphism predicts sleep structure and waking performance. Curr Biol 17: 613–618. [DOI] [PubMed] [Google Scholar]
- 15. Viola AU, James LM, Archer SN, Dijk DJ (2008) PER3 polymorphism and cardiac autonomic control: effects of sleep debt and circadian phase. Am J Physiol Heart Circ Physiol 295: H2156–H2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ellis J, von Schantz M, Jones KH, Archer SN (2009) Association between specific diurnal preference questionnaire items and PER3 VNTR genotype. Chronobiol Int 26: 464–473. [DOI] [PubMed] [Google Scholar]
- 17. Curtis DJ, Rasmussen MA (2006) The evolution of cathemerality in primates and other mammals: a comparative and chronoecological approach. Folia Primatol 77: 178–193. [DOI] [PubMed] [Google Scholar]
- 18.Ankel-Simons F, Rasmussen DT (2008) Diurnality, nocturnality, and the evolution of primate visual systems. Am J Phys Anthropol Suppl 47: 100–117. [DOI] [PubMed]
- 19. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kay RF (1997) Anthropoid origins. Science 275: 797–804. [DOI] [PubMed] [Google Scholar]
- 21. Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, et al. (2011) A molecular phylogeny of living primates. PLoS Genet 7: e1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Finnegan DJ (1989) Eukaryotic transposable elements and genome evolution. Trends Genet 5: 103–107. [DOI] [PubMed] [Google Scholar]
- 23. Kazazian HH Jr (2004) Mobile elements: drivers of genome evolution. Science 303: 1626–1632. [DOI] [PubMed] [Google Scholar]
- 24. Hedges DJ, Batzer MA (2005) From the margins of the genome: mobile elements shape primate evolution. Bioessays 27: 785–794. [DOI] [PubMed] [Google Scholar]
- 25. Medstrand P, van de Lagemaat LN, Dunn CA, Landry JR, Svenback D, et al. (2005) Impact of transposable elements on the evolution of mammalian gene regulation. Cytogenet Genome Res 110: 342–352. [DOI] [PubMed] [Google Scholar]
- 26.Eickbush TH, Eickbush DG (2006) Transposable elements: evolution. eSL.
- 27.Xing J, Witherspoon DJ, Ray DA, Batzer MA, Jorde LB (2007) Mobile DNA elements in primate and human evolution. Am J Phys Anthropol Suppl 45: 2–19. [DOI] [PubMed]
- 28. Cordaux R, Batzer MA (2009) The impact of retrotransposons on human genome evolution. Nat Rev Genet 10: 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ahmed M, Liang P (2012) Transposable elements are a significant contributor to tandem repeats in the human genome. Comp Funct Genomics 2012: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gemayel R, Vinces MD, Legendre M, Verstrepen KJ (2010) Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet 44: 445–477. [DOI] [PubMed] [Google Scholar]
- 31. Nakamura Y, Koyama K, Matsushima M (1998) VNTR (variable number of tandem repeat) sequences as transcriptional, translational, or functional regulators. J Hum Genet 43: 149–152. [DOI] [PubMed] [Google Scholar]
- 32. Fondon JW, Hammock EA, Hannan AJ, King DG (2008) Simple sequence repeats: genetic modulators of brain function and behavior. Trends Neurosci 31: 328–334. [DOI] [PubMed] [Google Scholar]
- 33. Matsushima N, Yoshida H, Kumaki Y, Kamiya M, Tanaka T, et al. (2008) Flexible structures and ligand interactions of tandem repeats consisting of proline, glycine, asparagine, serine, and/or threonine rich oligopeptides in proteins. Curr Protein Pept Sci 9: 591–610. [DOI] [PubMed] [Google Scholar]
- 34. Nithianantharajah J, Hannan AJ (2007) Dynamic mutations as digital genetic modulators of brain development, function and dysfunction. Bioessays 29: 525–535. [DOI] [PubMed] [Google Scholar]
- 35. Peixoto AA, Hennessy JM, Townson I, Hasan G, Rosbash M, et al. (1998) Molecular coevolution within a Drosophila clock gene. Proc Natl Acad Sci U S A 95: 4475–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sawyer LA, Sandrelli F, Pasetto C, Peixoto AA, Rosato E, et al. (2006) The period gene Thr-Gly polymorphism in Australian and African Drosophila melanogaster populations: implications for selection. Genetics 174: 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tobler I (1995) Is sleep fundamentally different between mammalian species? Behav Brain Res 69: 35–41. [DOI] [PubMed] [Google Scholar]
- 38. Fernandez-Duque E (2003) Influences of moonlight, ambient temperature, and food availability on the diurnal and nocturnal activity of owl monkeys (Aotus azarai). Behav Ecol Sociobiol 54: 431–440. [Google Scholar]
- 39. Heesy CP, Hall MI (2010) The nocturnal bottleneck and the evolution of mammalian vision. Brain Behav Evol 75: 195–203. [DOI] [PubMed] [Google Scholar]
- 40. Green P (2007) 2x genomes - does depth matter? Genome Res 17: 1547–1549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the PER3 VNTR regions from several primate species. Each repetition is represented in a different color. *Sequences obtained from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). **Sequences obtained from Jenkins et al. (2005).
(TIF)